Multi-Parameter Optimization in Directed Evolution: Engineering Thermostability, Enantioselectivity and Activity of an Epoxide Hydrolase
Li, G., Zhang, H., Sun, Z., Liu, X., Reetz, M.T.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| limonene epoxide hydrolase | 155 | Rhodococcus erythropolis | Mutation(s): 0 | 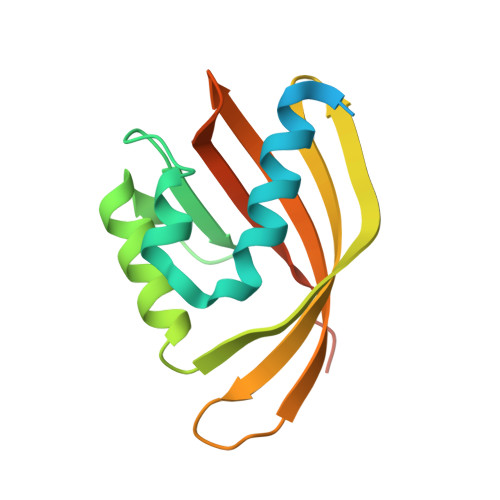 | |
UniProt | |||||
Find proteins for Q9ZAG3 (Rhodococcus erythropolis) Explore Q9ZAG3 Go to UniProtKB: Q9ZAG3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9ZAG3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 65.402 | α = 90 |
| b = 65.402 | β = 90 |
| c = 72.36 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Fundation of China | China | 31370925 |