Structural basis for potency differences between GDF8 and GDF11.
Walker, R.G., Czepnik, M., Goebel, E.J., McCoy, J.C., Vujic, A., Cho, M., Oh, J., Aykul, S., Walton, K.L., Schang, G., Bernard, D.J., Hinck, A.P., Harrison, C.A., Martinez-Hackert, E., Wagers, A.J., Lee, R.T., Thompson, T.B.(2017) BMC Biol 15: 19-19
- PubMed: 28257634
- DOI: https://doi.org/10.1186/s12915-017-0350-1
- Primary Citation of Related Structures:
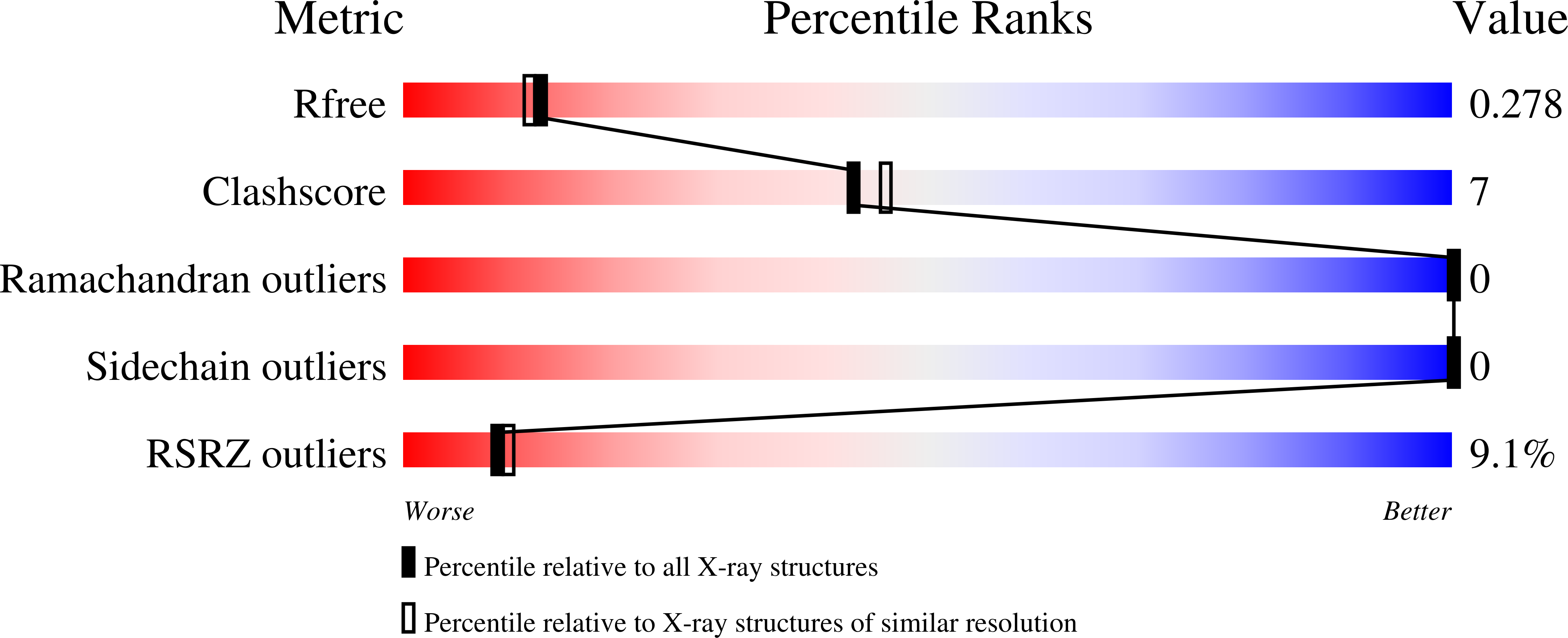
5JHW, 5JI1, 5UHM - PubMed Abstract:
Growth/differentiation factor 8 (GDF8) and GDF11 are two highly similar members of the transforming growth factor β (TGFβ) family. While GDF8 has been recognized as a negative regulator of muscle growth and differentiation, there are conflicting studies on the function of GDF11 and whether GDF11 has beneficial effects on age-related dysfunction. To address whether GDF8 and GDF11 are functionally identical, we compared their signaling and structural properties.
Organizational Affiliation:
Department of Molecular Genetics, Biochemistry, and Microbiology, University of Cincinnati, Cincinnati, OH, 45267, USA.