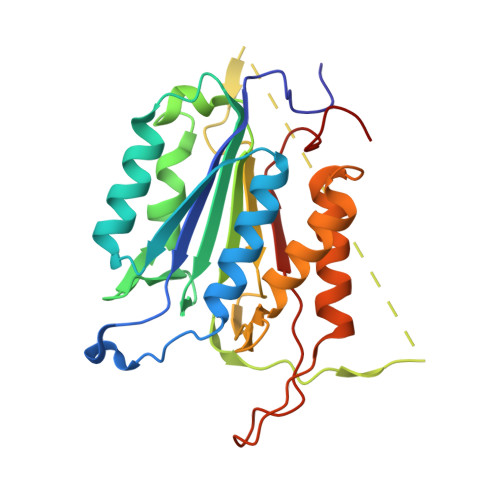
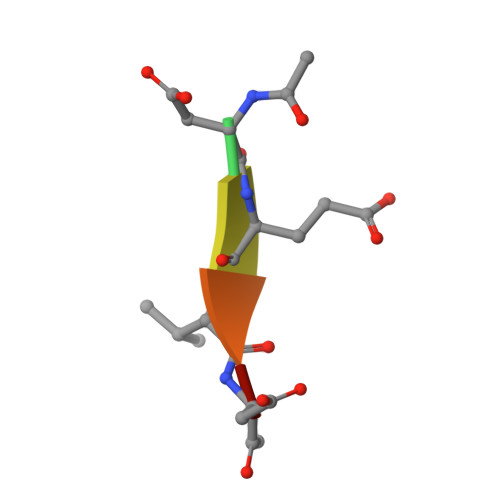
Phage display and structural studies reveal plasticity in substrate specificity of caspase-3a from zebrafish.
Tucker, M.B., MacKenzie, S.H., Maciag, J.J., Dirscherl Ackerman, H., Swartz, P., Yoder, J.A., Hamilton, P.T., Clay Clark, A.(2016) Protein Sci 25: 2076-2088
- PubMed: 27577093
- DOI: https://doi.org/10.1002/pro.3032
- Primary Citation of Related Structures:
5JFT - PubMed Abstract:
The regulation of caspase-3 enzyme activity is a vital process in cell fate decisions leading to cell differentiation and tissue development or to apoptosis. The zebrafish, Danio rerio, has become an increasingly popular animal model to study several human diseases because of their transparent embryos, short reproductive cycles, and ease of drug administration. While apoptosis is an evolutionarily conserved process in metazoans, little is known about caspases from zebrafish, particularly regarding substrate specificity and allosteric regulation compared to the human caspases. We cloned zebrafish caspase-3a (casp3a) and examined substrate specificity of the recombinant protein, Casp3a, compared to human caspase-3 (CASP3) by utilizing M13 bacteriophage substrate libraries that incorporated either random amino acids at P5-P1' or aspartate fixed at P1. The results show a preference for the tetrapeptide sequence DNLD for both enzymes, but the P4 position of zebrafish Casp3a also accommodates valine equally well. We determined the structure of zebrafish Casp3a to 2.28Å resolution by X-ray crystallography, and when combined with molecular dynamics simulations, the results suggest that a limited number of amino acid substitutions near the active site result in plasticity of the S4 sub-site by increasing flexibility of one active site loop and by affecting hydrogen-bonding with substrate. The data show that zebrafish Casp3a exhibits a broader substrate portfolio, suggesting overlap with the functions of caspase-6 in zebrafish development.
Organizational Affiliation:
Department of Molecular and Structural Biochemistry, NC State University, Raleigh, North Carolina, 27608.