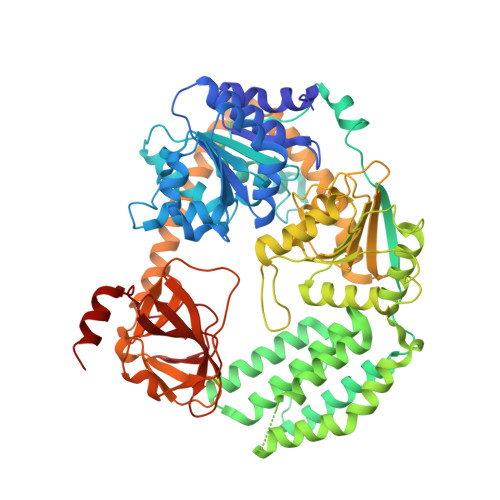
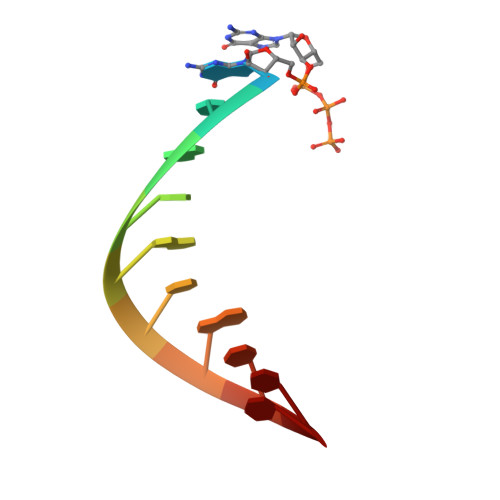
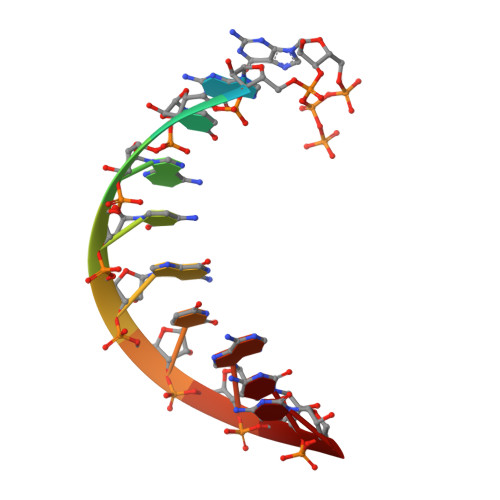
Structural Analysis of dsRNA Binding to Anti-viral Pattern Recognition Receptors LGP2 and MDA5.
Uchikawa, E., Lethier, M., Malet, H., Brunel, J., Gerlier, D., Cusack, S.(2016) Mol Cell 62: 586-602
- PubMed: 27203181
- DOI: https://doi.org/10.1016/j.molcel.2016.04.021
- Primary Citation of Related Structures:
5JAJ, 5JB2, 5JBG, 5JBJ, 5JC3, 5JC7, 5JCF, 5JCH - PubMed Abstract:
RIG-I and MDA5 sense virus-derived short 5'ppp blunt-ended or long dsRNA, respectively, causing interferon production. Non-signaling LGP2 appears to positively and negatively regulate MDA5 and RIG-I signaling, respectively. Co-crystal structures of chicken (ch) LGP2 with dsRNA display a fully or semi-closed conformation depending on the presence or absence of nucleotide. LGP2 caps blunt, 3' or 5' overhang dsRNA ends with 1 bp longer overall footprint than RIG-I. Structures of 1:1 and 2:1 complexes of chMDA5 with short dsRNA reveal head-to-head packing rather than the polar head-to-tail orientation described for long filaments. chLGP2 and chMDA5 make filaments with a similar axial repeat, although less co-operatively for chLGP2. Overall, LGP2 resembles a chimera combining a MDA5-like helicase domain and RIG-I like CTD supporting both stem and end binding. Functionally, RNA binding is required for LGP2-mediated enhancement of MDA5 activation. We propose that LGP2 end-binding may promote nucleation of MDA5 oligomerization on dsRNA.
Organizational Affiliation:
European Molecular Biology Laboratory, Grenoble Outstation, 71 Avenue des Martyrs, CS 90181, 38042 Grenoble Cedex 9, France; University Grenoble Alpes, Centre National de la Recherche Scientifique, EMBL Unit of Virus Host-Cell Interactions, 71 Avenue des Martyrs, CS 90181, 38042 Grenoble Cedex 9, France.