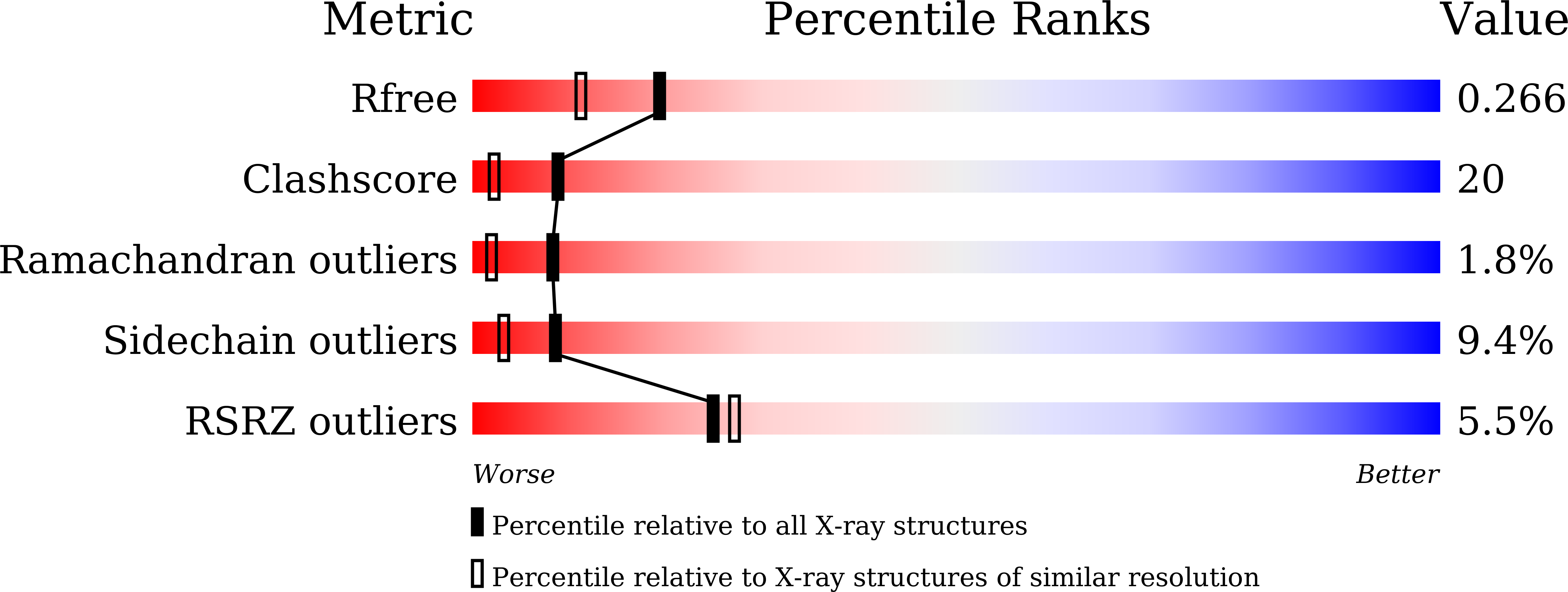
Crystal structure and substrate specificity of PTPN12.
Li, H., Yang, F., Liu, C., Xiao, P., Xu, Y.F., Liang, Z.L., Liu, C., Wang, H.M., Wang, W.J., Zheng, W.S., Zhang, W., Ma, X.Y., He, D.F., Song, X.Y., Cui, F.A., Xu, Z.G., Yi, F., Sun, J.P., Yu, X.(2016) Cell Rep 15: 1-14
- PubMed: 27134172
- DOI: https://doi.org/10.1016/j.celrep.2016.04.016
- Primary Citation of Related Structures:
5J8R - PubMed Abstract:
PTPN12 is an important tumor suppressor that plays critical roles in various physiological processes. However, the molecular basis underlying the substrate specificity of PTPN12 remains uncertain. Here, enzymological and crystallographic studies have enabled us to identify two distinct structural features that are crucial determinants of PTPN12 substrate specificity: the pY+1 site binding pocket and specific basic charged residues along its surface loops. Key structurally plastic regions and specific residues in PTPN12 enabled recognition of different HER2 phosphorylation sites and regulated specific PTPN12 functions. In addition, the structure of PTPN12 revealed a CDK2 phosphorylation site in a specific PTPN12 loop. Taken together, our results not only provide the working mechanisms of PTPN12 for desphosphorylation of its substrates but will also help in designing specific inhibitors of PTPN12.
Organizational Affiliation:
Key Laboratory Experimental Teratology of the Ministry of Education and Department of Physiology, Shandong University School of Medicine, 44 Wenhua Xi Road, Jinan, Shandong 250012, China; Department of Molecular Biology and Biochemistry, Shandong University School of Medicine, 44 Wenhua Xi Road, Jinan, Shandong 250012, China.