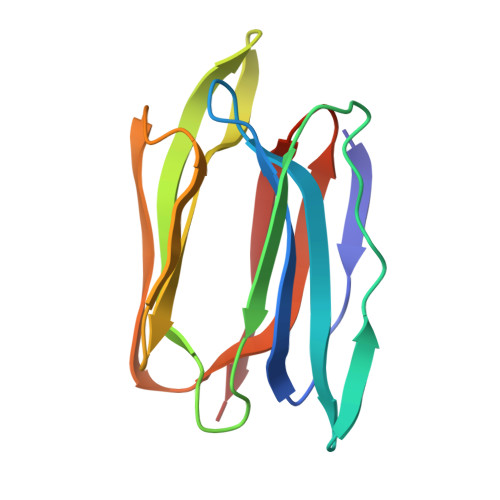
Distortion of the ligand molecule as a strategy for modulating binding affinity: Further studies involving complexes of jacalin with beta-substituted disaccharides.
Abhinav, K.V., Sharma, K., Surolia, A., Vijayan, M.(2017) IUBMB Life 69: 72-78
- PubMed: 28111895
- DOI: https://doi.org/10.1002/iub.1593
- Primary Citation of Related Structures:
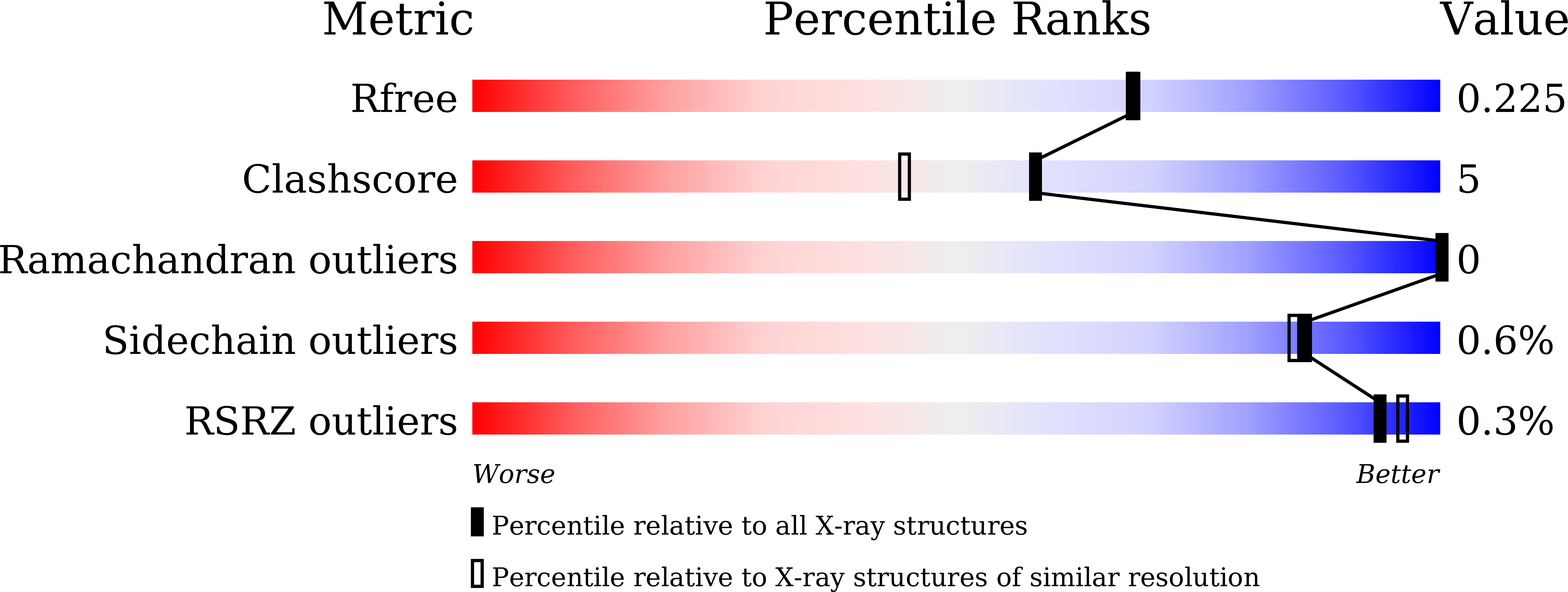
5J4T, 5J4X - PubMed Abstract:
Crystal structures of jacalin in complex with GlcNAc β-(1,3) Gal-β-OMe and Gal β-(1,3) Gal-β-OMe have been determined. The binding of the ligands to jacalin is similar to that of analogous α-substituted disaccharides. However, the β-substituted β-(1,3) linked disaccharides get distorted at the anomeric center and the glycosidic linkage. The distortion results in higher internal energies of the ligands leading to lower affinity to the lectin. This confirms the possibility of using ligand distortion as a strategy for modulating binding affinity. Unlike in the case of β-substituted monosaccharides bound to jacalin, where a larger distortion at the anomeric center was observed, smaller distortions are distributed among two centers in the structures of the two β-substituted β-(1,3) linked disaccharides presented here. These disaccharides, like the unsubstituted and α-substituted counterparts, bind jacalin with the reducing Gal at the primary binding site, indicating that the lower binding affinity of β-substituted disaccharides is not enough to overcome the intrinsic propensity of Gal β-(1,3) Gal-based disaccharides to bind jacalin with the reducing sugar at the primary site. © 2017 IUBMB Life, 69(2):72-78, 2017.
Organizational Affiliation:
Molecular Biophysics Unit, , Indian Institute of Science, Bangalore, India.