The binding specificity of RuTAP2dppz
Souter, J.E., Hall, J.P., Gurung, S.P., Brazier, J.A., Cardin, C.J.To be published.
Experimental Data Snapshot
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*CP*CP*GP*GP*GP*CP*CP*CP*GP*G)-3') | A [auth C] | 10 | synthetic construct | 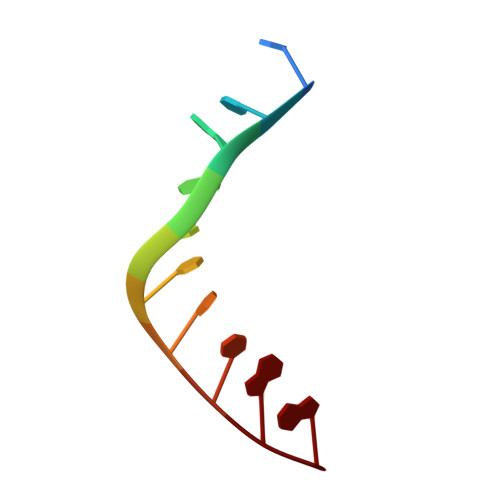 | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| RKL Query on RKL | C | Ru(tap)2(dppz) complex C38 H22 N12 Ru PIKVAZQLFXBUSD-UHFFFAOYSA-N |  | ||
| BA Query on BA | B [auth C] | BARIUM ION Ba XDFCIPNJCBUZJN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 46.783 | α = 90 |
| b = 46.783 | β = 90 |
| c = 34.948 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| SHELXDE | phasing |
| xia2 | data reduction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Biotechnology and Biological Sciences Research Council | United Kingdom | BB/K019279/1 |
| Biotechnology and Biological Sciences Research Council | United Kingdom | BB/M004635/1 |