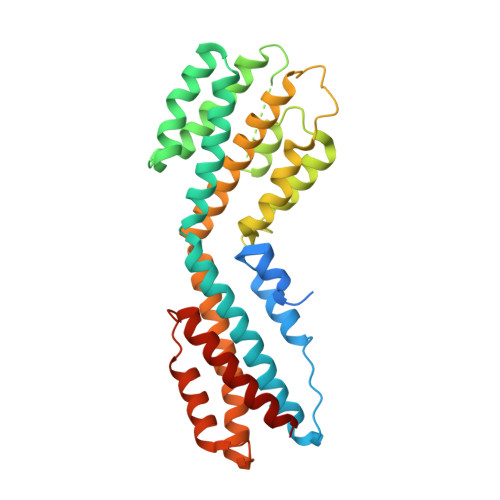
Structure of the Open Reading Frame 49 Protein Encoded by Kaposi's Sarcoma-Associated Herpesvirus.
Hew, K., Veerappan, S., Sim, D., Cornvik, T., Nordlund, P., Dahlroth, S.L.(2017) J Virol 91
- PubMed: 27807232
- DOI: https://doi.org/10.1128/JVI.01947-16
- Primary Citation of Related Structures:
5IPX - PubMed Abstract:
Herpesviruses alternate between the latent and the lytic life cycle. Switching into the lytic life cycle is important for herpesviral replication and disease pathogenesis. Activation of a transcription factor replication and transcription activator (RTA) has been demonstrated to govern this switch in Kaposi's sarcoma-associated herpesvirus (KSHV). The protein encoded by open reading frame 49 from KSHV (ORF49 KSHV ) has been shown to upregulate lytic replication in KSHV by enhancing the activities of the RTA. We have solved the crystal structure of the ORF49 KSHV protein to a resolution of 2.4 Å. The ORF49 KSHV protein has a novel fold consisting of 12 alpha-helices bundled into two pseudodomains. Most notably are distinct charged patches on the protein surface, which are possible protein-protein interaction sites. Homologs of the ORF49 KSHV protein in the gammaherpesvirus subfamily have low sequence similarities. Conserved residues are mainly located in the hydrophobic regions, suggesting that they are more likely to play important structural roles than functional ones. Based on the identification and position of three sulfates binding to the positive areas, we performed some initial protein-DNA binding studies by analyzing the thermal stabilization of the protein in the presence of DNA. The ORF49 KSHV protein is stabilized in a dose-responsive manner by double-stranded oligonucleotides, suggesting actual DNA interaction and binding. Biolayer interferometry studies also demonstrated that the ORF49 KSHV protein binds these oligonucleotides. Kaposi's sarcoma-associated herpesvirus (KSHV) is a tumorigenic gammaherpesvirus that causes multiple cancers and lymphoproliferative diseases. The virus exists mainly in the quiescent latent life cycle, but when it is reactivated into the lytic life cycle, new viruses are produced and disease symptoms usually manifest. Several KSHV proteins play important roles in this reactivation, but their exact roles are still largely unknown. In this study, we report the crystal structure of the open reading frame 49 protein encoded by KSHV (ORF49 KSHV ). Possible regions for protein interaction that could harbor functional importance were found on the surface of the ORF49 KSHV protein. This led to the discovery of novel DNA binding properties of the ORF49 KSHV protein. Evolutionary conserved structural elements with the functional homologs of ORF49 KSHV were also established with the structure.
Organizational Affiliation:
Division of Structural Biology and Biochemistry, Nanyang Technological University, School of Biological Sciences, Singapore.