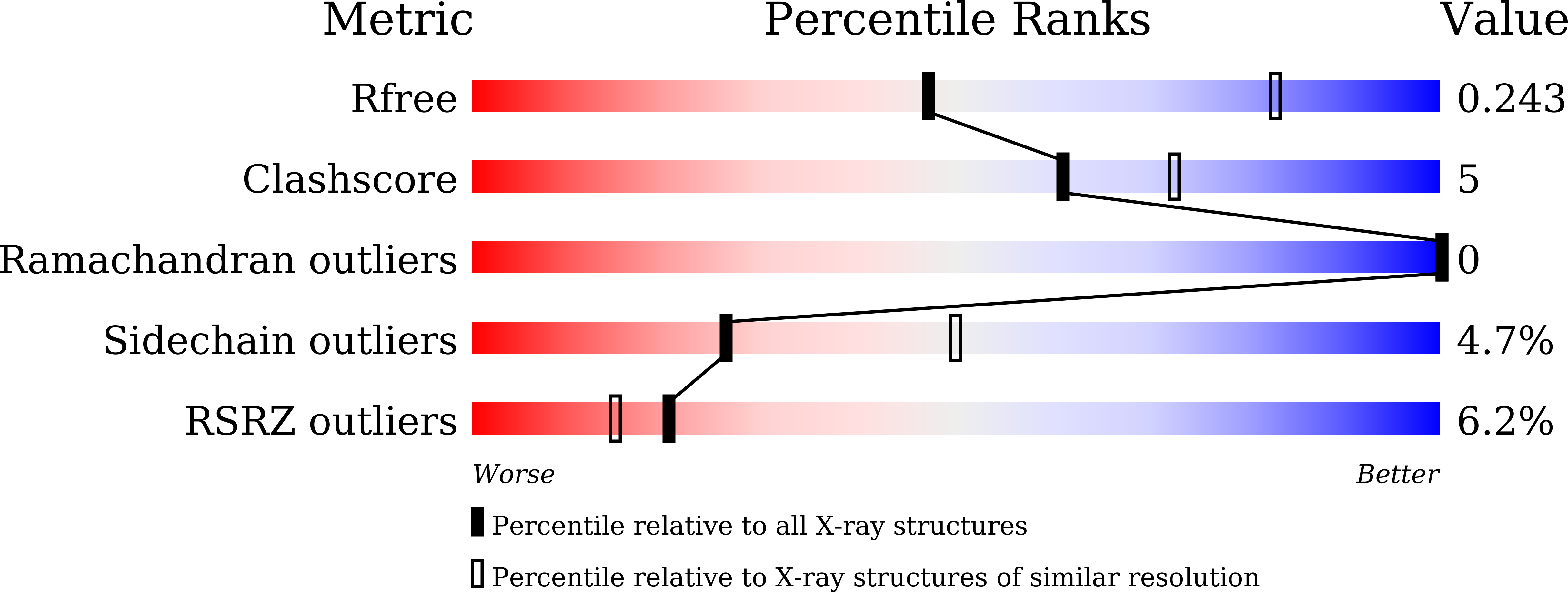
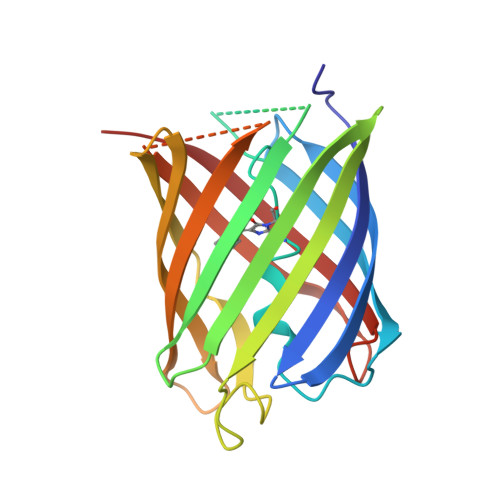
Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development
Kim, I.J., Kim, S., Park, J., Eom, I., Kim, S., Kim, J.H., Ha, S.C., Kim, Y.G., Hwang, K.Y., Nam, K.H.(2016) FEBS Lett 590: 2982-2990
- PubMed: 27433793
- DOI: https://doi.org/10.1002/1873-3468.12316
- Primary Citation of Related Structures:
5HZS, 5HZT, 5HZU - PubMed Abstract:
Many fluorescent proteins (FPs) show fluorescence quenching by specific metal ions, which can be applied towards metal biosensor development. In this study, we investigated the significant fluorescence quenching of Dronpa by Co(2+) and Cu(2+) ions. Crystal structures of Co(2+) -, Ni(2+) - and Cu(2+) -bound Dronpa revealed previously unseen, unique, metal-binding sites for fluorescence quenching. These metal ions commonly interact with surface-exposed histidine residues (His194-His210 and His210-His212), and interact indirectly with chromophores. Structural analysis of the Co(2+) - and Cu(2+) - binding sites of Dronpa provides insight into FP-based metal biosensor engineering.
Organizational Affiliation:
Division of Biotechnology, College of Life Sciences & Biotechnology, Korea University, Seoul, Korea.