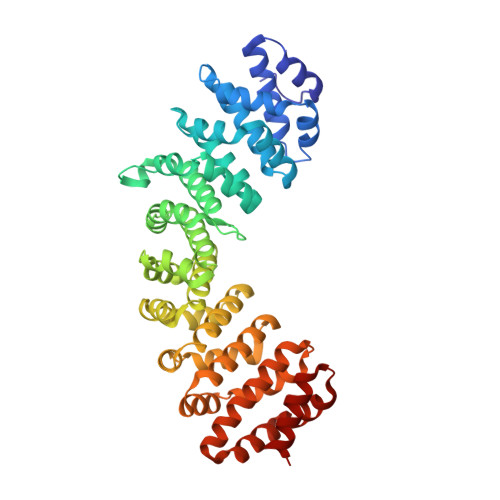
The C-terminal 18 Amino Acid Region of Dengue Virus NS5 Regulates its Subcellular Localization and Contains a Conserved Arginine Residue Essential for Infectious Virus Production.
Tay, M.Y., Smith, K., Ng, I.H., Chan, K.W., Zhao, Y., Ooi, E.E., Lescar, J., Luo, D., Jans, D.A., Forwood, J.K., Vasudevan, S.G.(2016) PLoS Pathog 12: e1005886-e1005886
- PubMed: 27622521
- DOI: https://doi.org/10.1371/journal.ppat.1005886
- Primary Citation of Related Structures:
5FC8, 5HHG - PubMed Abstract:
Dengue virus NS5 is the most highly conserved amongst the viral non-structural proteins and is responsible for capping, methylation and replication of the flavivirus RNA genome. Interactions of NS5 with host proteins also modulate host immune responses. Although replication occurs in the cytoplasm, an unusual characteristic of DENV2 NS5 is that it localizes to the nucleus during infection with no clear role in replication or pathogenesis. We examined NS5 of DENV1 and 2, which exhibit the most prominent difference in nuclear localization, employing a combination of functional and structural analyses. Extensive gene swapping between DENV1 and 2 NS5 identified that the C-terminal 18 residues (Cter18) alone was sufficient to direct the protein to the cytoplasm or nucleus, respectively. The low micromolar binding affinity between NS5 Cter18 and the nuclear import receptor importin-alpha (Impα), allowed their molecular complex to be purified, crystallised and visualized at 2.2 Å resolution using x-ray crystallography. Structure-guided mutational analysis of this region in GFP-NS5 clones of DENV1 or 2 and in a DENV2 infectious clone reveal residues important for NS5 subcellular localization. Notably, the trans conformation adopted by Pro-884 allows proper presentation for binding Impα and mutating this proline to Thr, as present in DENV1 NS5, results in mislocalizaion of NS5 to the cytoplasm without compromising virus fitness. In contrast, a single mutation to alanine at NS5 position R888, a residue conserved in all flaviviruses, resulted in a completely non-viable virus, and the R888K mutation led to a severely attenuated phentoype, even though NS5 was located in the nucleus. R888 forms a hydrogen bond with Y838 that is also conserved in all flaviviruses. Our data suggests an evolutionarily conserved function for NS5 Cter18, possibly in RNA interactions that are critical for replication, that is independent of its role in subcellular localization.
Organizational Affiliation:
Program in Emerging Infectious Diseases, Duke-NUS Medical School, Singapore.