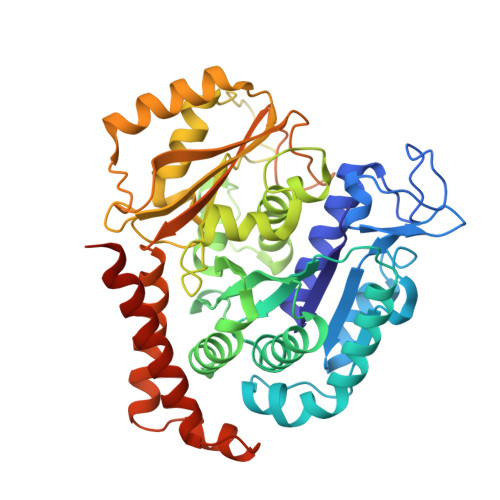
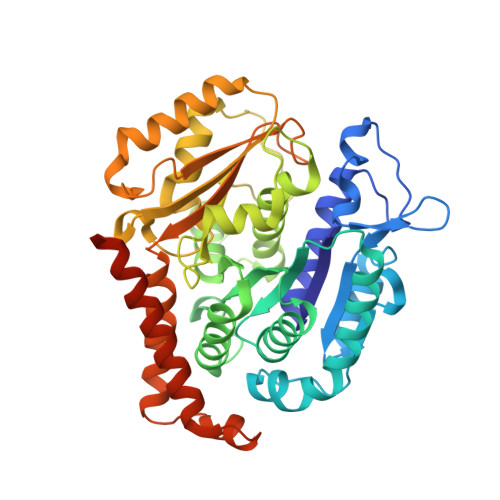
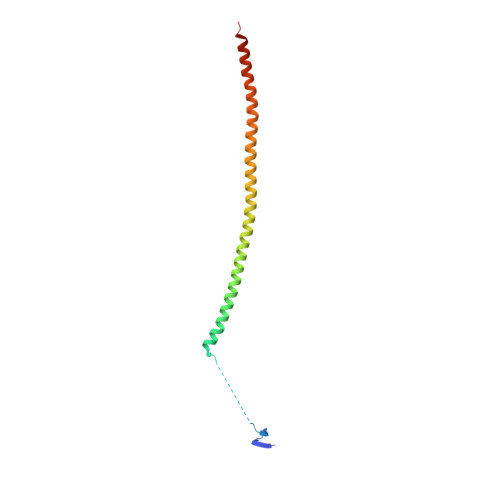
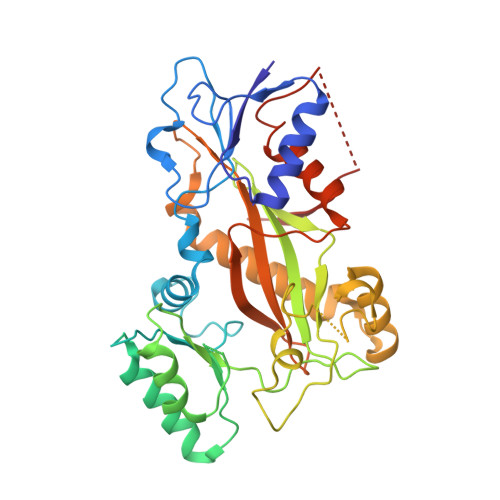
A Potent, Metabolically Stable Tubulin Inhibitor Targets the Colchicine Binding Site and Overcomes Taxane Resistance.
Arnst, K.E., Wang, Y., Hwang, D.J., Xue, Y., Costello, T., Hamilton, D., Chen, Q., Yang, J., Park, F., Dalton, J.T., Miller, D.D., Li, W.(2018) Cancer Res 78: 265-277
- PubMed: 29180476
- DOI: https://doi.org/10.1158/0008-5472.CAN-17-0577
- Primary Citation of Related Structures:
5H7O - PubMed Abstract:
Antimitotics that target tubulin are among the most useful chemotherapeutic drugs, but their clinical activity is often limited by the development of multidrug resistance. We recently discovered the novel small-molecule DJ101 as a potent and metabolically stable tubulin inhibitor that can circumvent the drug efflux pumps responsible for multidrug resistance of existing tubulin inhibitors. In this study, we determined the mechanism of action of this drug. The basis for its activity was illuminated by solving the crystal structure of DJ101 in complex with tubulin at a resolution of 2.8Å. Investigations of the potency of DJ101 in a panel of human metastatic melanoma cell lines harboring major clinically relevant mutations defined IC 50 values of 7-10 nmol/L. In cells, DJ101 disrupted microtubule networks, suppressed anchorage-dependent melanoma colony formation, and impaired cancer cell migration. In melanoma-bearing mice, DJ101 administration inhibited tumor growth and reduced lung metastasis in the absence of observable toxicity. DJ101 also completely inhibited tumor growth in a paclitaxel-resistant xenograft mouse model of human prostate cancer (PC-3/TxR), where paclitaxel was minimally effective. Our findings offer preclinical proof of concept for the continued development of DJ101 as a next-generation tubulin inhibitor for cancer therapy. Significance: These findings offer preclinical proof of concept for the continued development of DJ101 as a next-generation antitubulin drug for cancer therapy. Cancer Res; 78(1); 265-77. ©2017 AACR .
Organizational Affiliation:
Department of Pharmaceutical Sciences, College of Pharmacy, University of Tennessee Health Science Center, Memphis, Tennessee.