Structural insights into rice SalTol QTL located SALT protein.
Kaur, N., Sagar, A., Sharma, P., Pati, P.K.(2020) Sci Rep 10: 16589-16589
- PubMed: 33024209
- DOI: https://doi.org/10.1038/s41598-020-73517-y
- Primary Citation of Related Structures:
5GVY - PubMed Abstract:
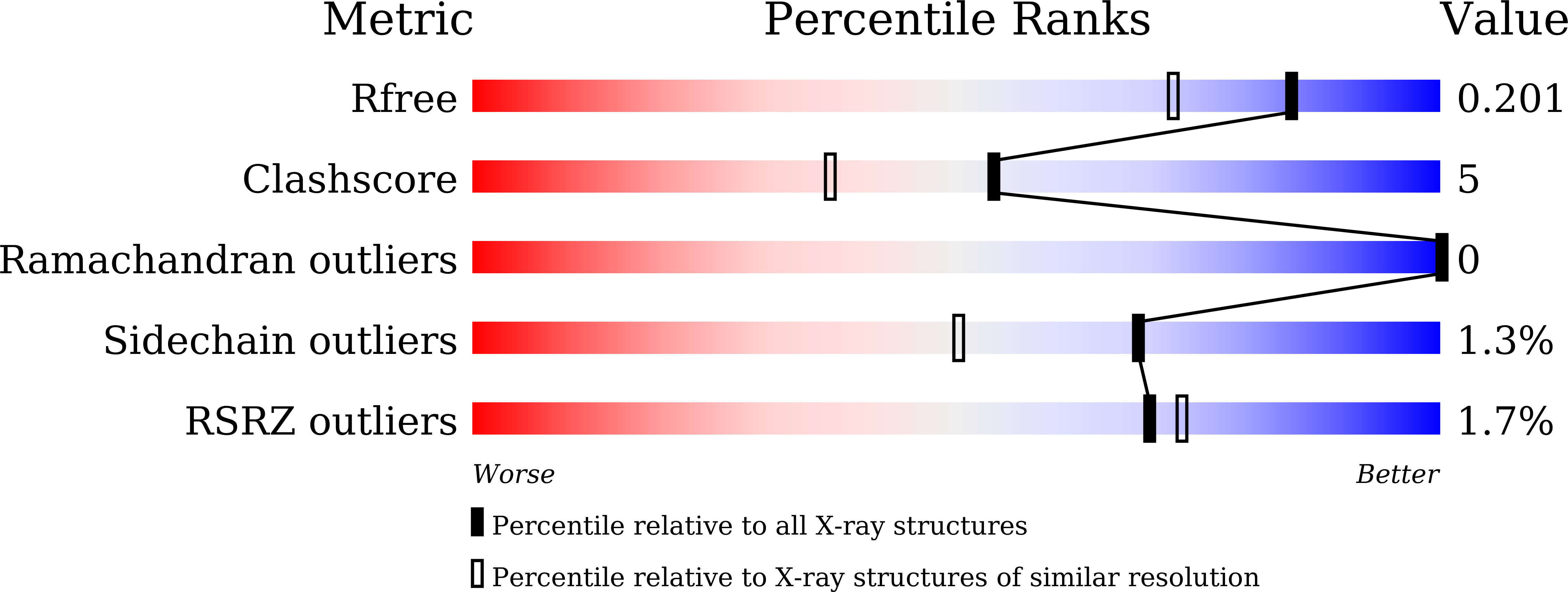
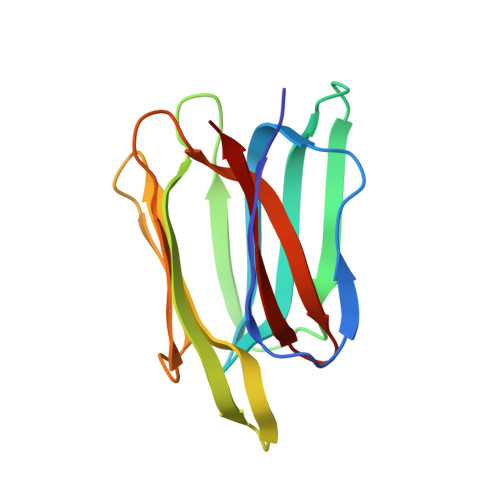
Salinity is one of the major stresses affecting rice production worldwide, and various strategies are being employed to increase salt tolerance. Recently, there has been resurgence of interest to characterize SalTol QTL harbouring number of critical genes involved in conferring salt stress tolerance in rice. The present study reports the structure of SALT, a SalTol QTL encoded protein by X-ray crystallography (PDB ID: 5GVY; resolution 1.66 Å). Each SALT chain was bound to one mannose via 8 hydrogen bonds. Compared to previous structure reported for similar protein, our structure showed a buried surface area of 900 Å 2 compared to only 240 Å 2 for previous one. Small-angle X-ray scattering (SAXS) data analysis showed that the predominant solution shape of SALT protein in solution is also dimer characterized by a radius of gyration and maximum linear dimension of 2.1 and 6.5 nm, respectively. The SAXS profiles and modelling confirmed that the dimeric association and relative positioning in solution matched better with our crystal structure instead of previously reported structure. Together, structural/biophysical data analysis uphold a tight dimeric structure for SALT protein with one mannose bound to each protein, which remains novel to date, as previous structures indicated one sugar unit sandwiched loosely between two protein chains.
Organizational Affiliation:
Department of Biotechnology, Guru Nanak Dev University, Amritsar, Punjab, 143005, India.