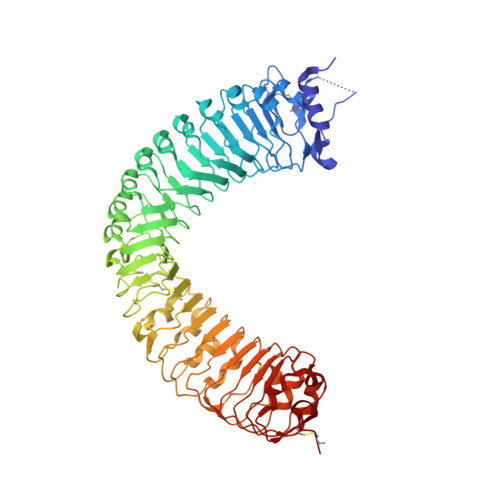
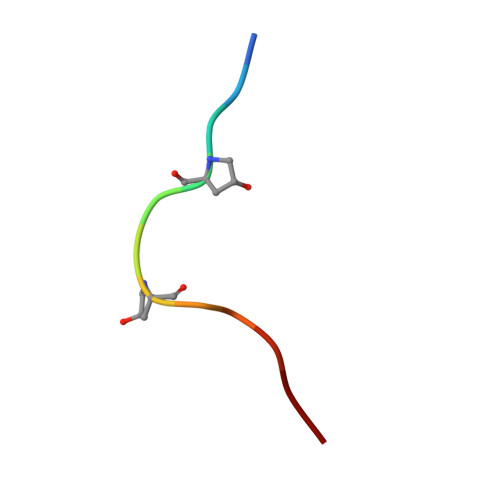
Crystal structure of PXY-TDIF complex reveals a conserved recognition mechanism among CLE peptide-receptor pairs
Zhang, H.Q., Lin, X.Y., Han, Z.F., Qu, L.J., Chai, J.J.(2016) Cell Res 26: 543-555
- PubMed: 27055373
- DOI: https://doi.org/10.1038/cr.2016.45
- Primary Citation of Related Structures:
5GR9 - PubMed Abstract:
Plants can achieve amazing lifespans because of their continuous and repetitive formation of new organs by stem cells present within meristems. The balance between proliferation and differentiation of meristem cells is largely regulated by the CLAVATA3/ENDOSPERM SURROUNDING REGION (CLE) peptide hormones. One of the well-characterized CLE peptides, CLE41/TDIF (tracheary elements differentiation inhibitory factor), functions to suppress tracheary element differentiation and promote procambial cell proliferation, playing important roles in vascular development and wood formation. The recognition mechanisms of TDIF or other CLE peptides by their respective receptors, however, remain largely elusive. Here we report the crystal structure of TDIF in complex with its receptor PXY, a leucine-rich repeat receptor kinase (LRR-RK). Our structure reveals that TDIF mainly adopts an "Ω"-like conformation binding to the inner surface of the LRR domain of PXY. Interaction between TDIF and PXY is predominately mediated by the relatively conserved amino acids of TDIF. Structure-based sequence alignment showed that the TDIF-interacting motifs are also conserved among other known CLE receptors. Our data provide a structural template for understanding the recognition mechanism of CLE peptides by their receptors, offering an opportunity for the identification of receptors of other uncharacterized CLE peptides.
Organizational Affiliation:
Ministry of Education Key Laboratory of Protein Science, Center for Structural Biology, School of Life Sciences, Tsinghua-Peking Joint Center for Life Sciences, Tsinghua University, Beijing 100084, China.