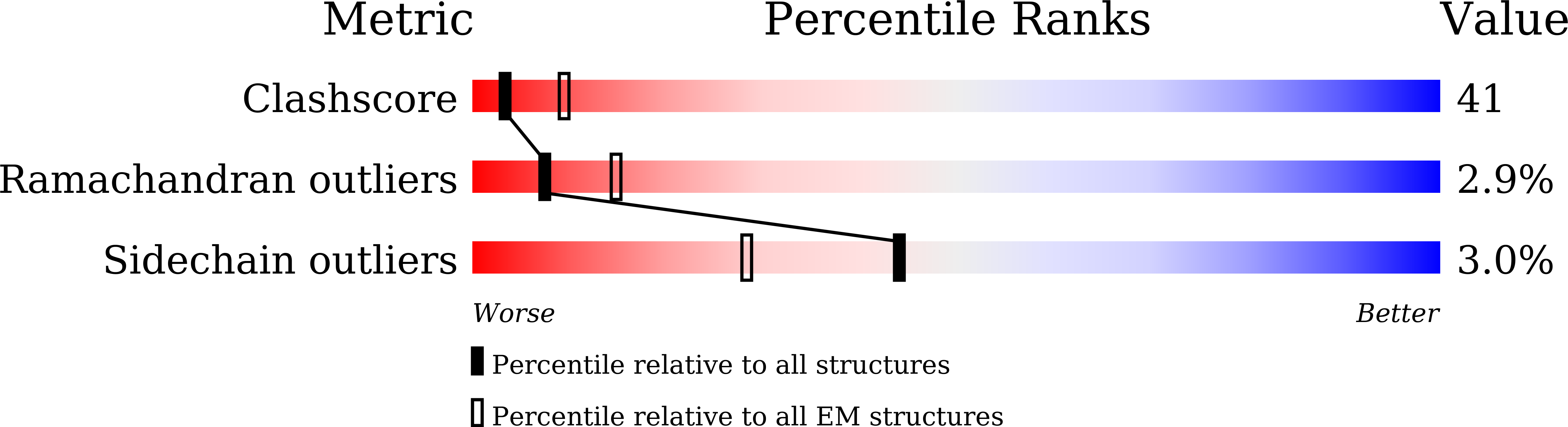Structure of human Aichi virus and implications for receptor binding
Zhu, L., Wang, X., Ren, J., Kotecha, A., Walter, T.S., Yuan, S., Yamashita, T., Tuthill, T.J., Fry, E.E., Rao, Z., Stuart, D.I.(2016) Nat Microbiol 1: 16150-16150
- PubMed: 27595320
- DOI: https://doi.org/10.1038/nmicrobiol.2016.150
- Primary Citation of Related Structures:
5GKA - PubMed Abstract:
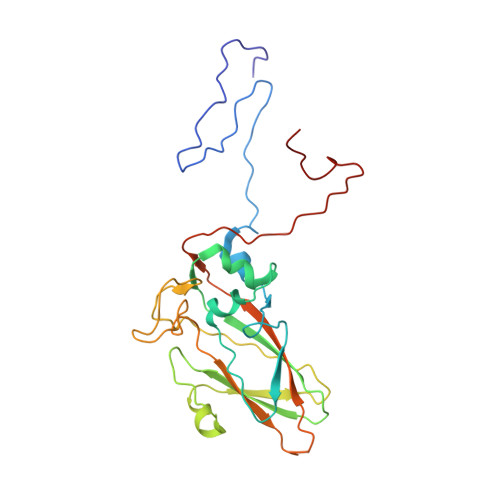
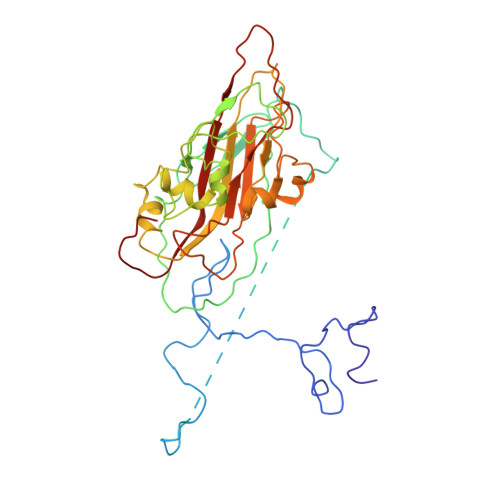
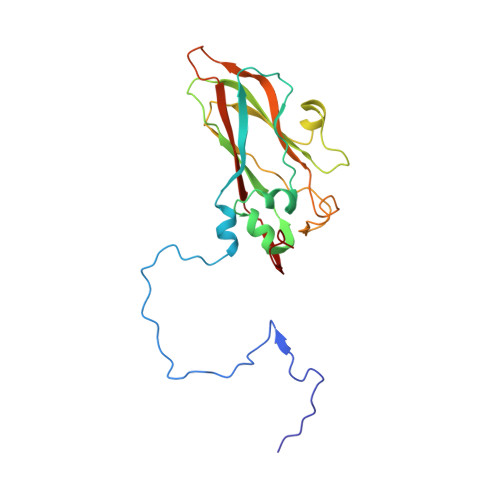
Aichi virus (AiV), an unusual and poorly characterized picornavirus, classified in the genus Kobuvirus, can cause severe gastroenteritis and deaths in children below the age of five years, especially in developing countries 1,2 . The seroprevalence of AiV is approximately 60% in children under the age of ten years and reaches 90% later in life 3,4 . There is no available vaccine or effective antiviral treatment. Here, we describe the structure of AiV at 3.7 Å. This first high-resolution structure for a kobuvirus is intermediate between those of the enteroviruses and cardioviruses, with a shallow, narrow depression bounded by the prominent VP0 CD loops (linking the C and D strands of the β-barrel), replacing the depression known as the canyon, frequently the site of receptor attachment in enteroviruses. VP0 is not cleaved to form VP2 and VP4, so the 'VP2' β-barrel structure is complemented with a unique extended structure on the inside of the capsid. On the outer surface, a polyproline helix structure, not seen previously in picornaviruses is present at the C terminus of VP1, a position where integrin binding motifs are found in some other picornaviruses. A peptide corresponding to this polyproline motif somewhat attenuates virus infectivity, presumably blocking host-cell attachment. This may guide cellular receptor identification.
Organizational Affiliation:
Division of Structural Biology, University of Oxford, The Henry Wellcome Building for Genomic Medicine, Headington, Oxford OX3 7BN, UK.