Novel Interaction Mechanism of a Domain Antibody Based Inhibitor of Human Vascular Endothelial Growth Factor with Greater Potency Than Ranibizumab and Bevacizumab and Improved Capacity Over Aflibercept.
Walker, A., Chung, C., Neu, M., Burman, M., Batuwangala, T., Jones, G., Tang, C., Steward, M., Mullin, M., Tournier, N., Lewis, A., Korczynska, J., Chung, V., Catchpole, I.(2016) J Biol Chem 291: 5500
- PubMed: 26728464
- DOI: https://doi.org/10.1074/jbc.M115.691162
- Primary Citation of Related Structures:
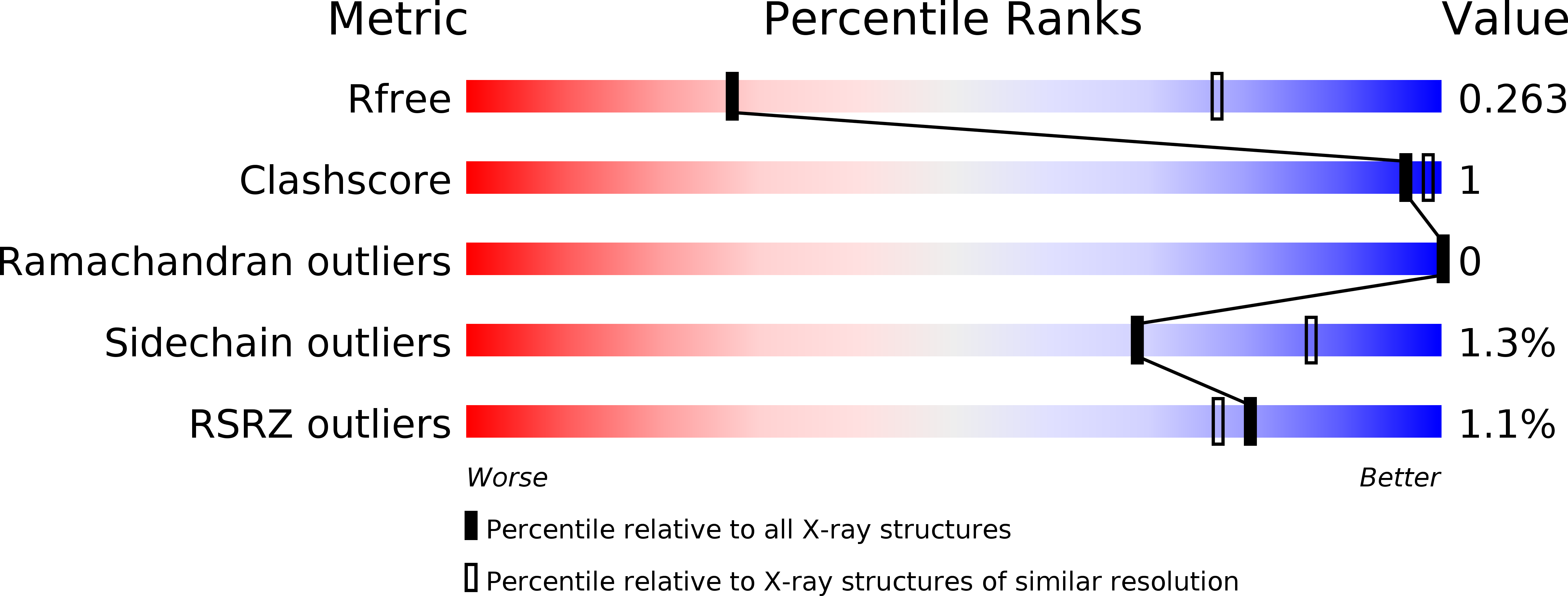
5FV1, 5FV2 - PubMed Abstract:
A potent VEGF inhibitor with novel antibody architecture and antigen binding mode has been developed. The molecule, hereafter referred to as VEGF dual dAb (domain antibody), was evaluated in vitro for binding to VEGF and for potency in VEGF-driven models and compared with other anti-VEGF biologics that have been used in ocular anti-angiogenic therapeutic regimes. VEGF dual dAb is more potent than bevacizumab and ranibizumab for VEGF binding, inhibition of VEGF receptor binding assays (RBAs), and VEGF-driven in vitro models of angiogenesis and displays comparable inhibition to aflibercept (Eylea). VEGF dual dAb is dimeric, and each monomer contains two distinct anti-VEGF domain antibodies attached via linkers to a human IgG1 Fc domain. Mechanistically, the enhanced in vitro potency of VEGF dual dAb, in comparison to other anti-VEGF biologics, can be explained by increased binding stoichiometry. A consistent model of the target engagement has been built based on the x-ray complexes of each of the two isolated domain antibodies with the VEGF antigen.
Organizational Affiliation:
From BioPharm Innovation.