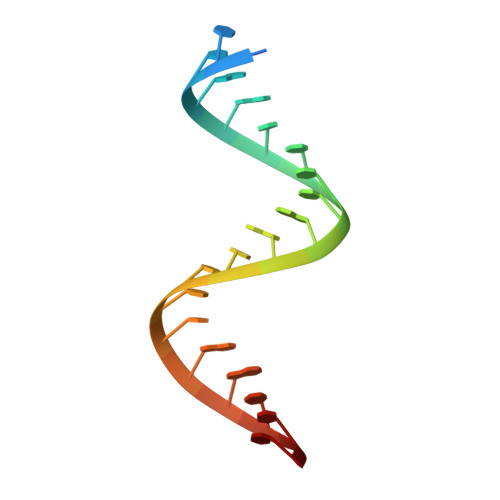
Pathogenic C9ORF72 Antisense Repeat RNA Forms a Double Helix with Tandem C:C Mismatches.
Dodd, D.W., Tomchick, D.R., Corey, D.R., Gagnon, K.T.(2016) Biochemistry 55: 1283-1286
- PubMed: 26878348
- DOI: https://doi.org/10.1021/acs.biochem.6b00136
- Primary Citation of Related Structures:
5EW4, 5EW7 - PubMed Abstract:
Expansion of a GGGGCC/CCCCGG repeat sequence in the first intron of the C9ORF72 gene is a leading cause of frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS). In this combined disorder, called c9FTD/ALS, the expansion is bidirectionally transcribed into sense and antisense repeat RNA associated with disease. To better understand the role of C9ORF72 repeat RNA in molecular disease pathology, we determined crystal structures of a [(CCCCGG)3(CCCC)] model antisense repeat RNA to 1.47 Å resolution. The RNA structure was an A-form-like double helix composed of repeating and regularly spaced tandem C:C mismatch pairs that perturbed helical geometry and surface charge. Solution studies revealed a preference for A-form-like helical conformations as the repeat number increased. Results provide a structural starting point for rationalizing the contribution of repeat RNA to c9FTD/ALS molecular disease mechanisms and for developing molecules to target C9ORF72 repeat RNA as potential therapeutics.
Organizational Affiliation:
Department of Pharmacology, ‡Department of Biophysics, and §Department of Biochemistry, University of Texas Southwestern Medical Center , Dallas, Texas 75390, United States.