The Discovery and Characterization of K-756, a Novel Wnt/ beta-Catenin Pathway Inhibitor Targeting Tankyrase
Okada-Iwasaki, R., Takahashi, Y., Watanabe, Y., Ishida, H., Saito, J., Nakai, R., Asai, A.(2016) Mol Cancer Ther 15: 1525-1534
- PubMed: 27196752
- DOI: https://doi.org/10.1158/1535-7163.MCT-15-0938
- Primary Citation of Related Structures:
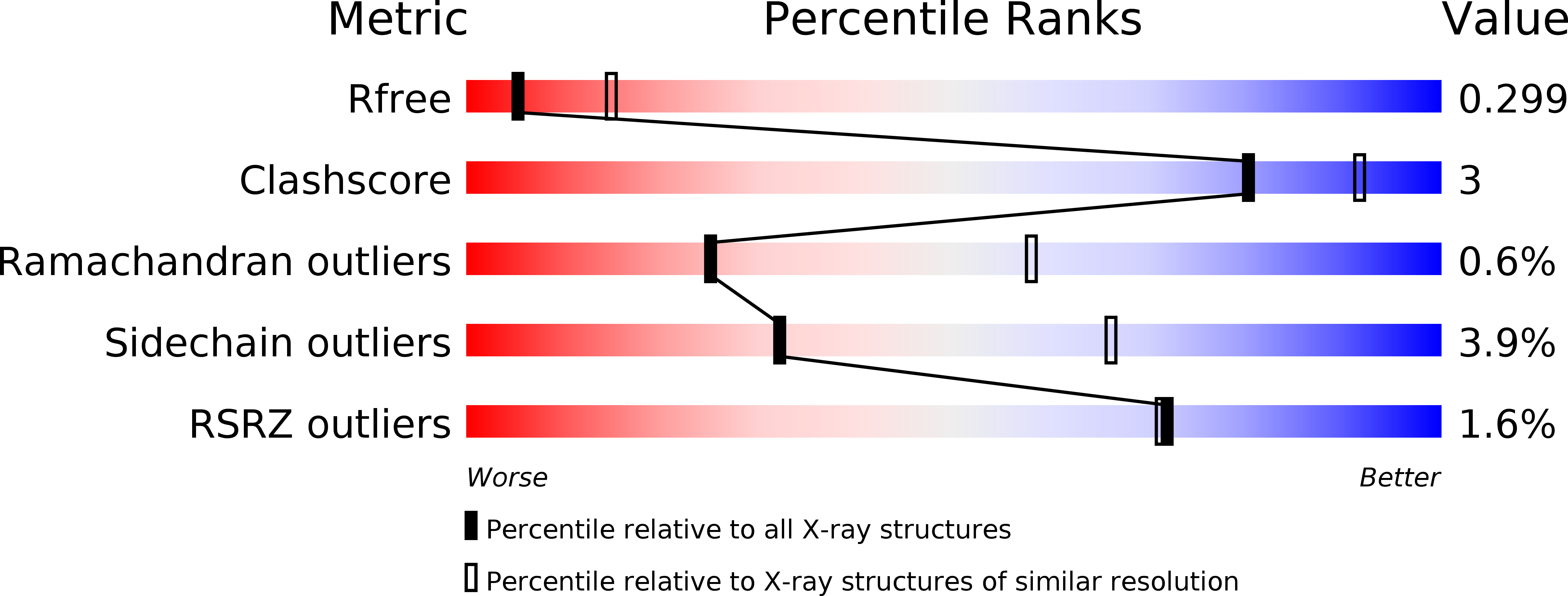
5ETY - PubMed Abstract:
The Wnt/β-catenin pathway is a well-known oncogenic pathway. Its suppression has long been considered as an important challenge in treating cancer patients. Among colon cancer patients in particular, most patients carry an adenomatous polyposis coli (APC) mutation that leads to an aberration of Wnt/β-catenin pathway. To discover the small molecule inhibitors of the Wnt/β-catenin pathway, we conducted high-throughput screening in APC-mutant colon cancer DLD-1 cells using a transcriptional reporter assay, which identified a selective Wnt/β-catenin pathway inhibitor, K-756. K-756 stabilizes Axin and reduces active β-catenin, and inhibits the genes downstream of endogenous Wnt/β-catenin. We subsequently identified that K-756 is a tankyrase (TNKS) inhibitor. TNKS, which belongs to the PARP family, poly-ADP ribosylates Axin and promotes Axin degradation via the proteasome pathway. K-756 binds to the induced pocket of TNKS and inhibits its enzyme activity. Moreover, PARP family enzyme assays showed that K-756 is a selective TNKS inhibitor. K-756 inhibited the cell growth of APC-mutant colorectal cancer COLO 320DM and SW403 cells by inhibiting the Wnt/β-catenin pathway. An in vivo study showed that the oral administration of K-756 inhibited the Wnt/β-catenin pathway in colon cancer xenografts in mice. To further explore the therapeutic potential of K-756, we also evaluated the effects of K-756 in non-small cell lung cancer cells. Although a single treatment of K-756 did not induce antiproliferative activity, when K-756 was combined with an EGFR inhibitor (gefitinib), it showed a strong synergistic effect. Therefore, K-756, a novel selective TNKS inhibitor, could be a leading compound in the development of anticancer agents. Mol Cancer Ther; 15(7); 1525-34. ©2016 AACR.
Organizational Affiliation:
R&D Division, Kyowa Hakko Kirin Co., Ltd., Shizuoka, Japan. Center for Drug Discovery, Graduate School of Pharmaceutical Sciences, University of Shizuoka, Shizuoka, Japan.