Crystal Structure and Biochemical Characterization of a Mycobacterium smegmatis AAA-Type Nucleoside Triphosphatase Phosphohydrolase (Msm0858).
Unciuleac, M.C., Smith, P.C., Shuman, S.(2016) J Bacteriol 198: 1521-1533
- PubMed: 26953339
- DOI: https://doi.org/10.1128/JB.00905-15
- Primary Citation of Related Structures:
5E7P - PubMed Abstract:
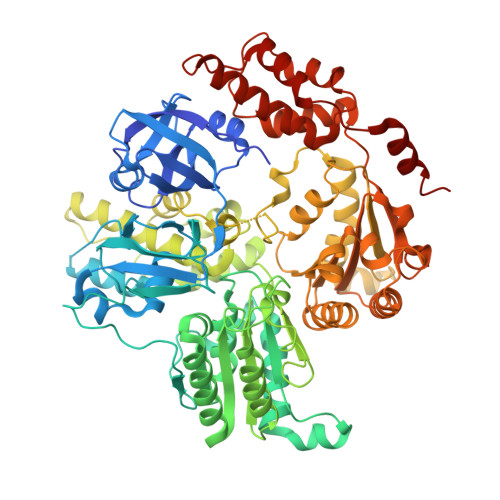
AAA proteins (ATPases associated with various cellular activities) use the energy of ATP hydrolysis to drive conformational changes in diverse macromolecular targets. Here, we report the biochemical characterization and 2.5-Å crystal structure of a Mycobacterium smegmatis AAA protein Msm0858, the ortholog of Mycobacterium tuberculosis Rv0435c. Msm0858 is a magnesium-dependent ATPase and is active with all nucleoside triphosphates (NTPs) and deoxynucleoside triphosphates (dNTPs) as substrates. The Msm0858 structure comprises (i) an N-terminal domain (amino acids [aa] 17 to 201) composed of two β-barrel modules and (ii) two AAA domains, D1 (aa 212 to 473) and D2 (aa 476 to 744), each of which has ADP in the active site. Msm0858-ADP is a monomer in solution and in crystallized form. Msm0858 domains are structurally homologous to the corresponding modules of mammalian p97. However, the position of the N-domain modules relative to the AAA domains in the Msm0858-ADP tertiary structure is different and would impede the formation of a p97-like hexameric quaternary structure. Mutational analysis of the A-box and B-box motifs indicated that the D1 and D2 AAA domains are both capable of ATP hydrolysis. Simultaneous mutations of the D1 and D2 active-site motifs were required to abolish ATPase activity. ATPase activity was effaced by mutation of the putative D2 arginine finger, suggesting that Msm0858 might oligomerize during the ATPase reaction cycle. A truncated variant Msm0858 (aa 212 to 745) that lacks the N domain was characterized as a catalytically active homodimer. Recent studies have underscored the importance of AAA proteins (ATPases associated with various cellular activities) in the physiology of mycobacteria. This study reports the ATPase activity and crystal structure of a previously uncharacterized mycobacterial AAA protein, Msm0858. Msm0858 consists of an N-terminal β-barrel domain and two AAA domains, each with ADP bound in the active site. Msm0858 is a structural homolog of mammalian p97, with respect to the linear order and tertiary structures of their domains.
Organizational Affiliation:
Molecular Biology Program, Sloan-Kettering Institute, New York, New York, USA.