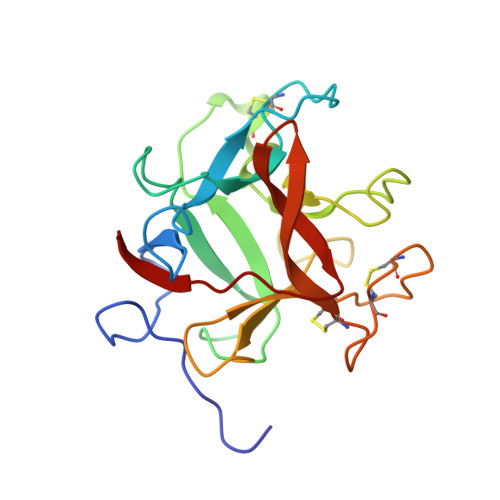
Structure of a Kunitz-type potato cathepsin D inhibitor.
Guo, J., Erskine, P.T., Coker, A.R., Wood, S.P., Cooper, J.B.(2015) J Struct Biol 192: 554-560
- PubMed: 26542926
- DOI: https://doi.org/10.1016/j.jsb.2015.10.020
- Primary Citation of Related Structures:
5DZU - PubMed Abstract:
Potato cathepsin D inhibitor (PDI) is a glycoprotein of 188 amino acids which can inhibit both the aspartic protease cathepsin D and the serine protease trypsin. Here we report the first X-ray structure of PDI at a resolution of 2.1 Å showing that PDI adopts a β-trefoil fold, which is typical of the Kunitz-family protease inhibitors, with the inhibitory loops protruding from the core. Possible reactive-site loops including one involving a unique disulphide and another involving a protruding 310 helix are identified and docking studies indicate the mode of action of this unusual bi-functional inhibitor.
Organizational Affiliation:
Division of Medicine, UCL, Gower Street, London WC1E 6BT, United Kingdom.