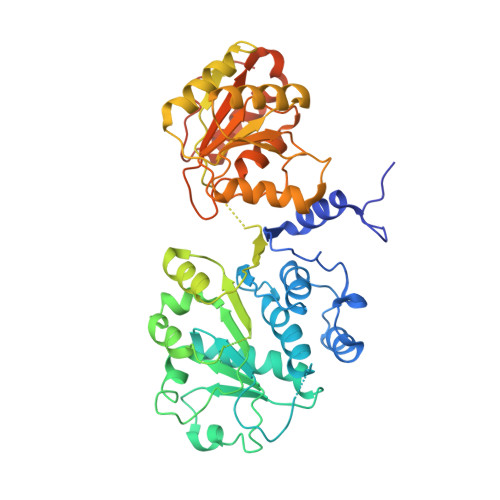
Structural analysis of the spliceosomal RNA helicase Prp28 from the thermophilic eukaryote Chaetomium thermophilum.
Tauchert, M.J., Ficner, R.(2016) Acta Crystallogr F Struct Biol Commun 72: 409-416
- PubMed: 27139834
- DOI: https://doi.org/10.1107/S2053230X16006038
- Primary Citation of Related Structures:
5DTU - PubMed Abstract:
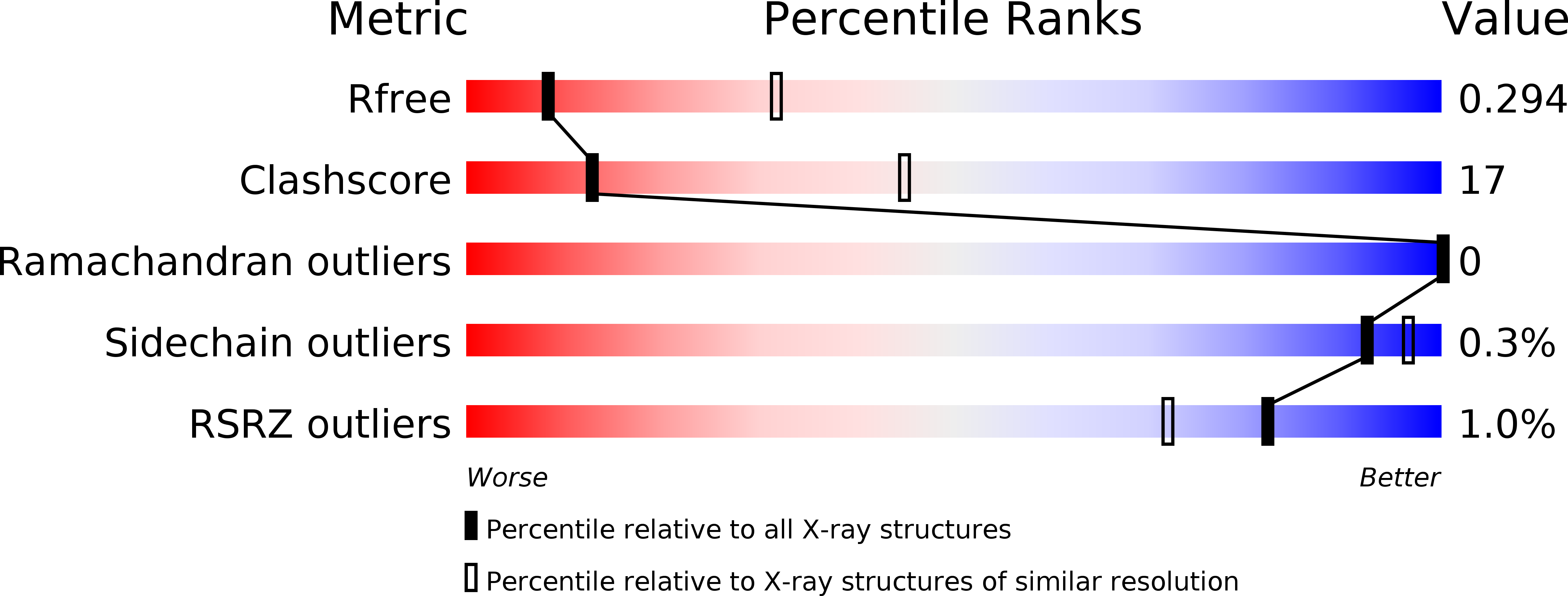
Prp28 (pre-mRNA-splicing ATP-dependent RNA helicase 28) is a spliceosomal DEAD-box helicase which is involved in two steps of spliceosome assembly. It is required for the formation of commitment complex 2 in an ATP-independent manner as well as for the formation of the pre-catalytic spliceosome, which in contrast is ATP-dependent. During the latter step, Prp28 is crucial for the integration of the U4/U6·U5 tri-snRNP since it displaces the U1 snRNP and allows the U6 snRNP to base-pair with the 5'-splice site. Here, the crystal structure of Prp28 from the thermophilic fungus Chaetomium thermophilum is reported at 3.2 Å resolution and is compared with the available structures of homologues.
Organizational Affiliation:
Department of Molecular Structural Biology, Institute for Microbiology and Genetics, GZMB, Georg-August-University Göttingen, Justus-von-Liebig Weg 11, 37077 Göttingen, Germany.