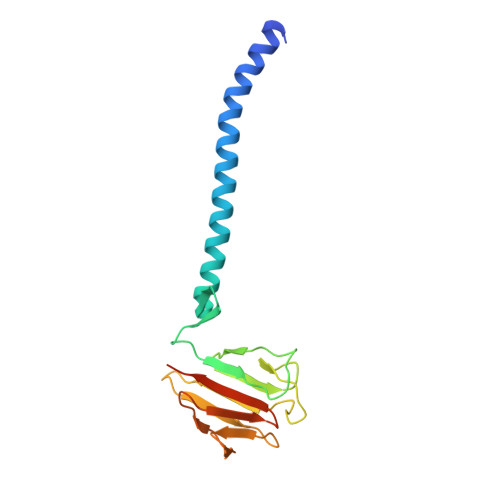
Structural Correlation of the Neck Coil with the Coiled-coil (CC1)-Forkhead-associated (FHA) Tandem for Active Kinesin-3 KIF13A
Ren, J., Huo, L., Wang, W., Zhang, Y., Li, W., Lou, J., Xu, T., Feng, W.(2016) J Biol Chem 291: 3581-3594
- PubMed: 26680000
- DOI: https://doi.org/10.1074/jbc.M115.689091
- Primary Citation of Related Structures:
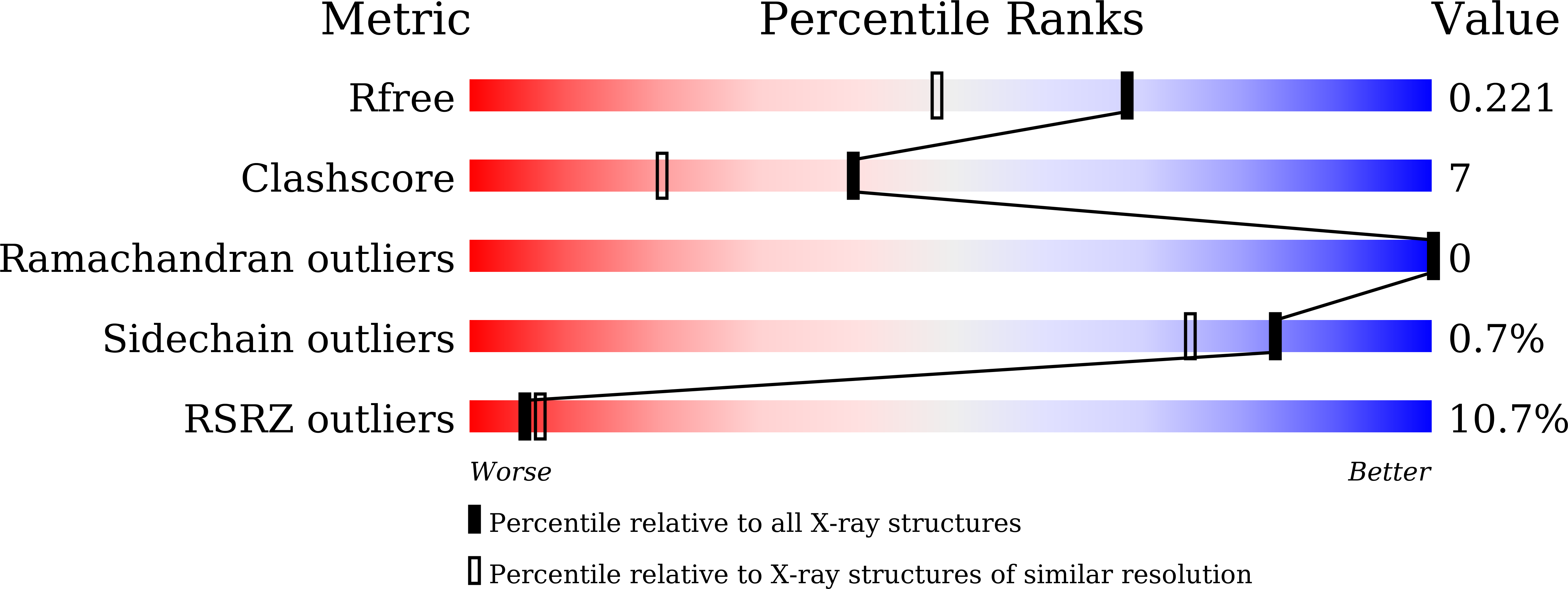
5DJN, 5DJO - PubMed Abstract:
Processive kinesin motors often contain a coiled-coil neck that controls the directionality and processivity. However, the neck coil (NC) of kinesin-3 is too short to form a stable coiled-coil dimer. Here, we found that the coiled-coil (CC1)-forkhead-associated (FHA) tandem (that is connected to NC by Pro-390) of kinesin-3 KIF13A assembles as an extended dimer. With the removal of Pro-390, the NC-CC1 tandem of KIF13A unexpectedly forms a continuous coiled-coil dimer that can be well aligned into the CC1-FHA dimer. The reverse introduction of Pro-390 breaks the NC-CC1 coiled-coil dimer but provides the intrinsic flexibility to couple NC with the CC1-FHA tandem. Mutations of either NC, CC1, or the FHA domain all significantly impaired the motor activity. Thus, the three elements within the NC-CC1-FHA tandem of KIF13A are structurally interrelated to form a stable dimer for activating the motor. This work also provides the first direct structural evidence to support the formation of a coiled-coil neck by the short characteristic neck domain of kinesin-3.
Organizational Affiliation:
From the National Laboratory of Biomacromolecules and.