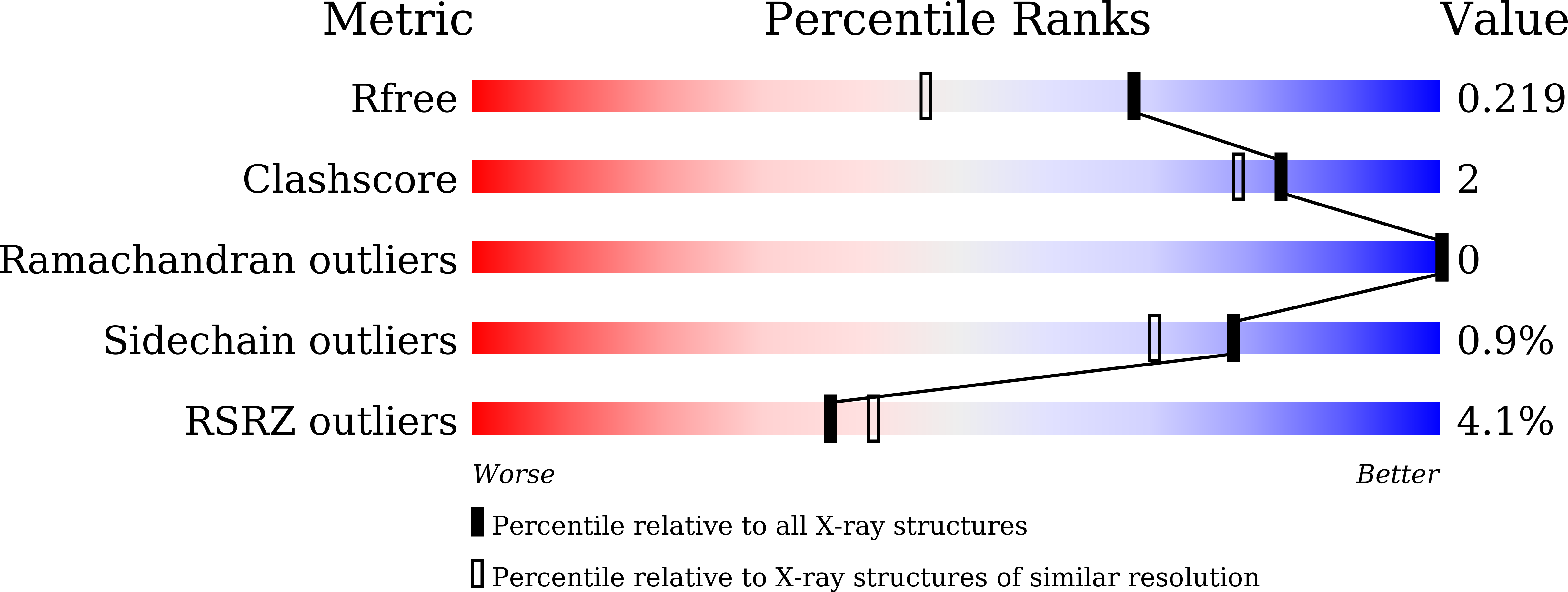
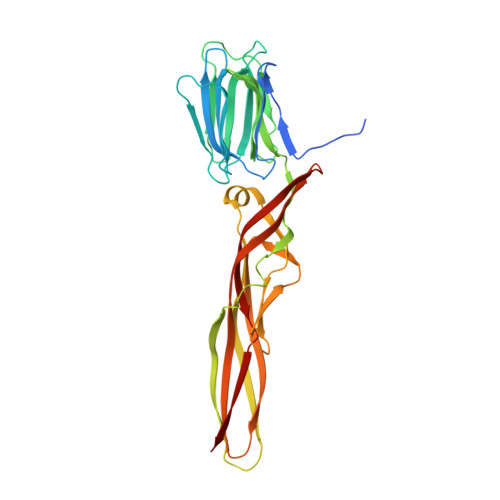
Structural basis for receptor recognition and pore formation of a zebrafish aerolysin-like protein.
Jia, N., Liu, N., Cheng, W., Jiang, Y.L., Sun, H., Chen, L.L., Peng, J., Zhang, Y., Ding, Y.H., Zhang, Z.H., Wang, X., Cai, G., Wang, J., Dong, M.Q., Zhang, Z., Wu, H., Wang, H.W., Chen, Y., Zhou, C.Z.(2016) EMBO Rep 17: 235-248
- PubMed: 26711430
- DOI: https://doi.org/10.15252/embr.201540851
- Primary Citation of Related Structures:
4ZNO, 4ZNQ, 4ZNR, 5DI0 - PubMed Abstract:
Various aerolysin-like pore-forming proteins have been identified from bacteria to vertebrates. However, the mechanism of receptor recognition and/or pore formation of the eukaryotic members remains unknown. Here, we present the first crystal and electron microscopy structures of a vertebrate aerolysin-like protein from Danio rerio, termed Dln1, before and after pore formation. Each subunit of Dln1 dimer comprises a β-prism lectin module followed by an aerolysin module. Specific binding of the lectin module toward high-mannose glycans triggers drastic conformational changes of the aerolysin module in a pH-dependent manner, ultimately resulting in the formation of a membrane-bound octameric pore. Structural analyses combined with computational simulations and biochemical assays suggest a pore-forming process with an activation mechanism distinct from the previously characterized bacterial members. Moreover, Dln1 and its homologs are ubiquitously distributed in bony fishes and lamprey, suggesting a novel fish-specific defense molecule.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at the Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, China.