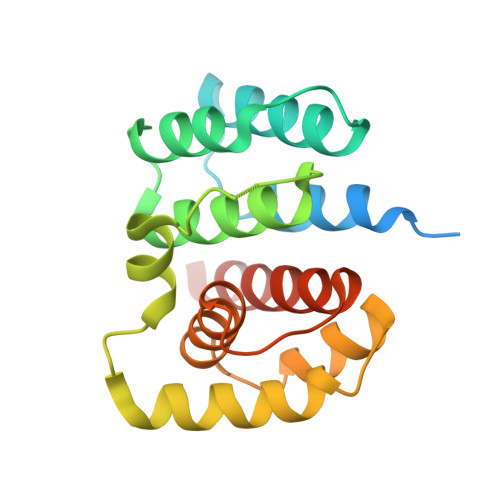
2.1 Angstrom resolution crystal structure of matrix protein 1 (M1; residues 1-164) from Influenza A virus (A/Puerto Rico/8/34(H1N1))
Halavaty, A.S., Minasov, G., Flores, K., Dubrovska, I., Grimshaw, S., Shuvalova, L., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.