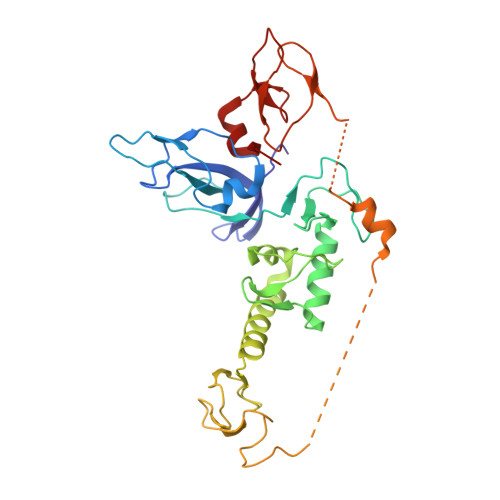
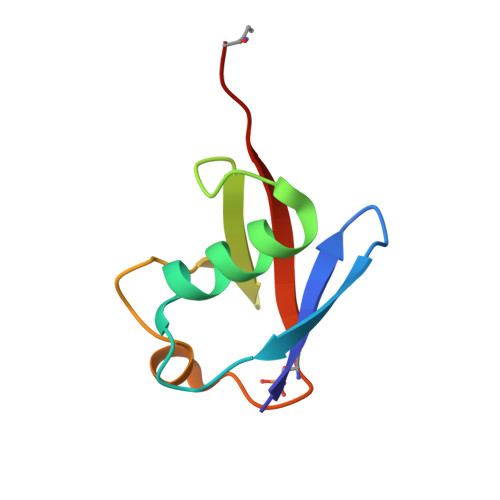
Mechanism of phospho-ubiquitin-induced PARKIN activation.
Wauer, T., Simicek, M., Schubert, A., Komander, D.(2015) Nature 524: 370-374
- PubMed: 26161729
- DOI: https://doi.org/10.1038/nature14879
- Primary Citation of Related Structures:
5C9V, 5CAW - PubMed Abstract:
The E3 ubiquitin ligase PARKIN (encoded by PARK2) and the protein kinase PINK1 (encoded by PARK6) are mutated in autosomal-recessive juvenile Parkinsonism (AR-JP) and work together in the disposal of damaged mitochondria by mitophagy. PINK1 is stabilized on the outside of depolarized mitochondria and phosphorylates polyubiquitin as well as the PARKIN ubiquitin-like (Ubl) domain. These phosphorylation events lead to PARKIN recruitment to mitochondria, and activation by an unknown allosteric mechanism. Here we present the crystal structure of Pediculus humanus PARKIN in complex with Ser65-phosphorylated ubiquitin (phosphoUb), revealing the molecular basis for PARKIN recruitment and activation. The phosphoUb binding site on PARKIN comprises a conserved phosphate pocket and harbours residues mutated in patients with AR-JP. PhosphoUb binding leads to straightening of a helix in the RING1 domain, and the resulting conformational changes release the Ubl domain from the PARKIN core; this activates PARKIN. Moreover, phosphoUb-mediated Ubl release enhances Ubl phosphorylation by PINK1, leading to conformational changes within the Ubl domain and stabilization of an open, active conformation of PARKIN. We redefine the role of the Ubl domain not only as an inhibitory but also as an activating element that is restrained in inactive PARKIN and released by phosphoUb. Our work opens up new avenues to identify small-molecule PARKIN activators.
Organizational Affiliation:
Medical Research Council Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge CB2 0QH, UK.