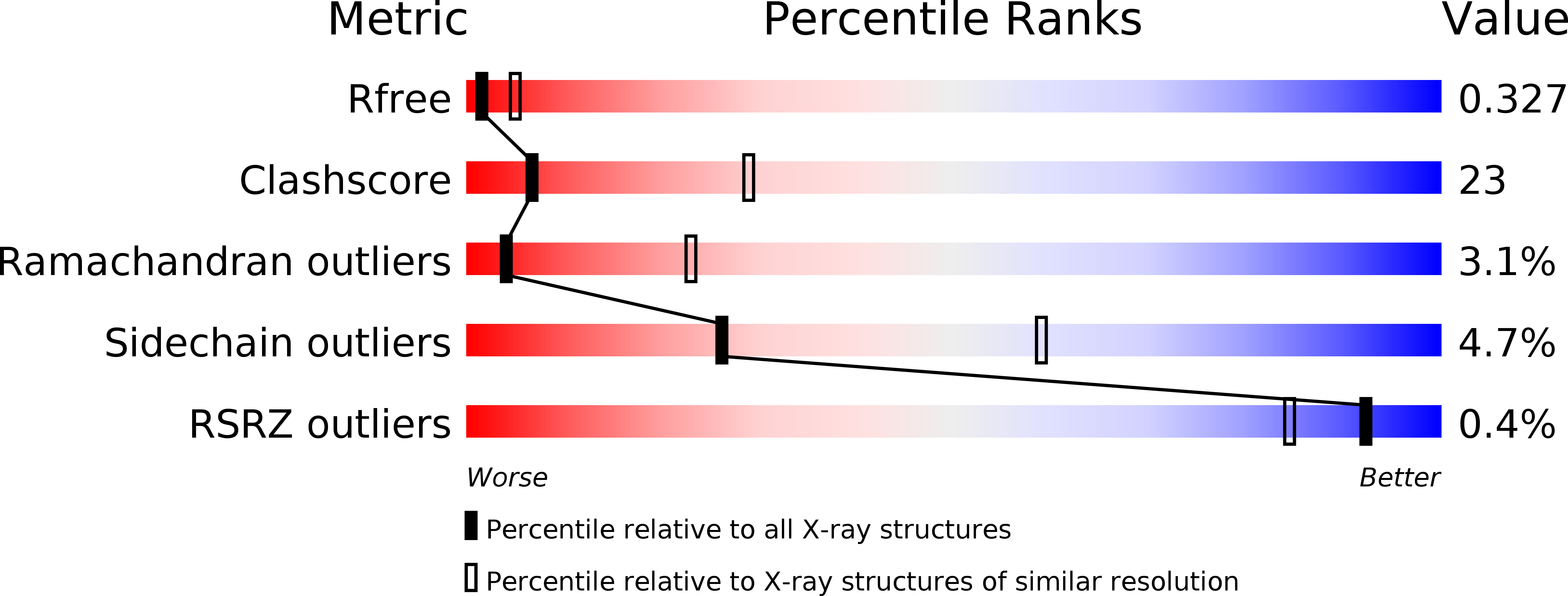
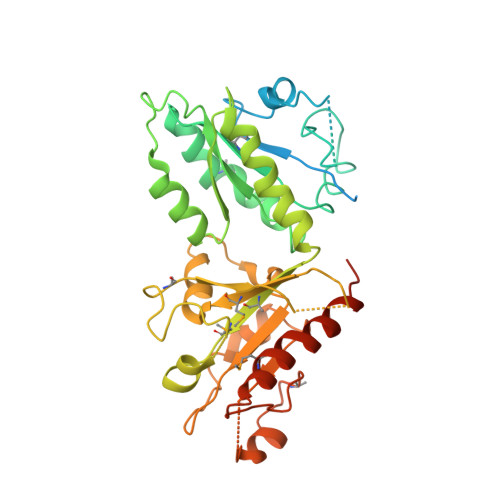
Crystal Structure Analysis of Wild Type and Fast Hydrolyzing Mutant of EhRabX3, a Tandem Ras Superfamily GTPase from Entamoeba histolytica.
Srivastava, V.K., Chandra, M., Saito-Nakano, Y., Nozaki, T., Datta, S.(2016) J Mol Biol 428: 41-51
- PubMed: 26555751
- DOI: https://doi.org/10.1016/j.jmb.2015.11.003
- Primary Citation of Related Structures:
5C1S, 5C1T - PubMed Abstract:
The enteric protozoan parasite, Entamoeba histolytica, is the causative agent of amoebic dysentery, liver abscess and colitis in human. Vesicular trafficking plays a key role in the survival and virulence of the protozoan and is regulated by various Rab GTPases. EhRabX3 is a catalytically inefficient amoebic Rab protein, which is unique among the eukaryotic Ras superfamily by virtue of its tandem domain organization. Here, we report the crystal structures of GDP-bound fast hydrolyzing mutant (V71A/K73Q) and GTP-bound wild type EhRabX3 at 3.1 and 2.8Å resolutions, respectively. Though both G-domains possess "phosphate binding loop containing nucleoside triphosphate hydrolases fold", only the N-terminal domain binds to guanine nucleotide. The relative orientation of the N-terminal domain and C-terminal domain is stabilized by numerous inter-domain interactions. Compared to other Ras superfamily members, both the GTPase domains displayed large deviation in switch II perhaps due to non-conservative substitutions in this region. As a result, entire switch II is restructured and moved away from the nucleotide binding pocket, providing a rationale for the diminished GTPase activity of EhRabX3. The N-terminal GTPase domain possesses unusually large number of cysteine residues. X-ray crystal structure of the fast hydrolyzing mutant of EhRabX3 revealed that C39 and C163 formed an intra-molecular disulfide bond. Subsequent mutational and biochemical studies suggest that C39 and C163 are critical for maintaining the structural integrity and function of EhRabX3. Structure-guided functional investigation of cysteine mutants could provide the physiological implications of the disulfide bond and could allow us to design potential inhibitors for the better treatment of intestinal amebiasis.
Organizational Affiliation:
Department of Biological Sciences, Indian Institute of Science Education and Research Bhopal 462023, India.