Structural characterization and modeling of the Borrelia burgdorferi hybrid histidine kinase Hk1 periplasmic sensor: A system for sensing small molecules associated with tick feeding.
Bauer, W.J., Luthra, A., Zhu, G., Radolf, J.D., Malkowski, M.G., Caimano, M.J.(2015) J Struct Biol 192: 48-58
- PubMed: 26321039
- DOI: https://doi.org/10.1016/j.jsb.2015.08.013
- Primary Citation of Related Structures:
5BWJ - PubMed Abstract:
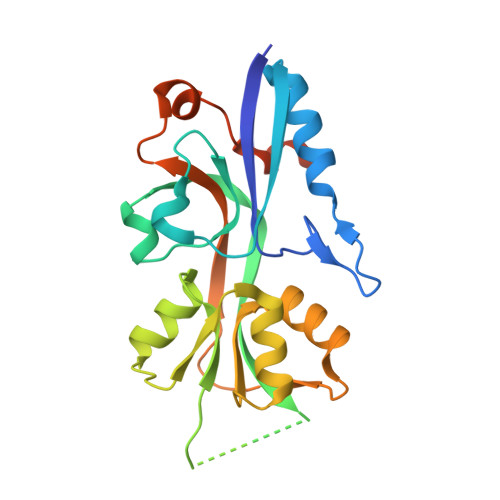
Two-component signal transduction systems are the primary mechanisms by which bacteria perceive and respond to changes in their environment. The Hk1/Rrp1 two-component system (TCS) in Borrelia burgdorferi consists of a hybrid histidine kinase and a response regulator with diguanylate cyclase activity, respectively. Phosphorylated Rrp1 catalyzes the synthesis of c-di-GMP, a second messenger associated with bacterial life-style control networks. Spirochetes lacking either Hk1 or Rrp1 are virulent in mice but destroyed within feeding ticks. Activation of Hk1 by exogenous stimuli represents the seminal event for c-di-GMP signaling. We reasoned that structural characterization of Hk1's sensor would provide insights into the mechanism underlying signal transduction and aid in the identification of activating ligands. The Hk1 sensor is composed of three ligand-binding domains (D1-3), each with homology to periplasmic solute-binding proteins (PBPs) typically associated with ABC transporters. Herein, we determined the structure for D1, the most N-terminal PBP domain. As expected, D1 displays a bilobed Venus Fly Trap-fold. Similar to the prototypical sensor PBPs HK29S from Geobacter sulfurreducens and VFT2 from Bordetella pertussis, apo-D1 adopts a closed conformation. Using complementary approaches, including SAXS, we established that D1 forms a dimer in solution. The D1 structure enabled us to model the D2 and D3 domains. Differences in the ligand-binding pockets suggest that each PBP recognizes a different ligand. The ability of Hk1 to recognize multiple stimuli provides spirochetes with a means of distinguishing between the acquisition and transmission blood meals and generate a graded output response that is reflective of the perceived environmental threats.
Organizational Affiliation:
Hauptman-Woodward Medical Research Institute, Buffalo, NY 14203.