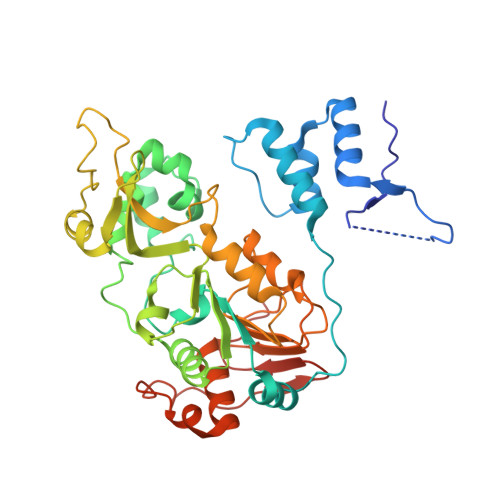
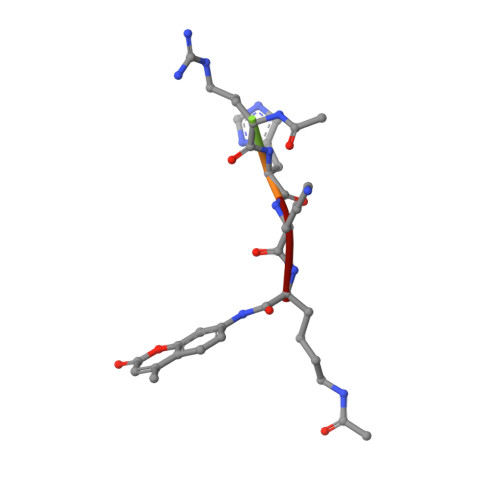
Structural basis for allosteric, substrate-dependent stimulation of SIRT1 activity by resveratrol
Cao, D., Wang, M., Qiu, X., Liu, D., Jiang, H., Yang, N., Xu, R.M.(2015) Genes Dev 29: 1316-1325
- PubMed: 26109052
- DOI: https://doi.org/10.1101/gad.265462.115
- Primary Citation of Related Structures:
5BTR - PubMed Abstract:
Sirtuins with an extended N-terminal domain (NTD), represented by yeast Sir2 and human SIRT1, harbor intrinsic mechanisms for regulation of their NAD-dependent deacetylase activities. Elucidation of the regulatory mechanisms is crucial for understanding the biological functions of sirtuins and development of potential therapeutics. In particular, SIRT1 has emerged as an attractive therapeutic target, and the search for SIRT1-activating compounds (STACs) has been actively pursued. However, the effectiveness of a class of reported STACs (represented by resveratrol) as direct SIRT1 activators is under debate due to the complication involving the use of fluorogenic substrates in in vitro assays. Future efforts of SIRT1-based therapeutics necessitate the dissection of the molecular mechanism of SIRT1 stimulation. We solved the structure of SIRT1 in complex with resveratrol and a 7-amino-4-methylcoumarin (AMC)-containing peptide. The structure reveals the presence of three resveratrol molecules, two of which mediate the interaction between the AMC peptide and the NTD of SIRT1. The two NTD-bound resveratrol molecules are principally responsible for promoting tighter binding between SIRT1 and the peptide and the stimulation of SIRT1 activity. The structural information provides valuable insights into regulation of SIRT1 activity and should benefit the development of authentic SIRT1 activators.
Organizational Affiliation:
National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China;