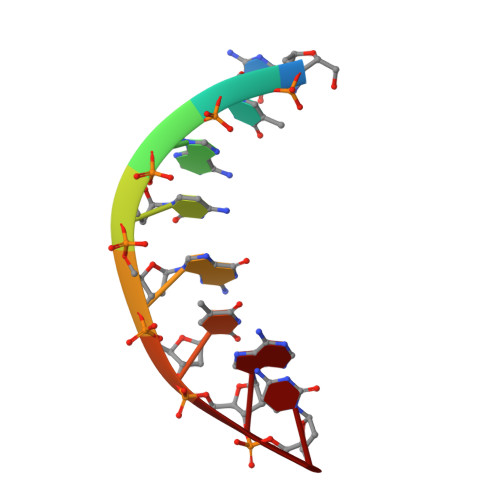
The crystal structure of d(GTACGTAC) at 2.25 A resolution: are the A-DNA's always unwound approximately 10 degrees at the C-G steps
Takusagawa, F.(1990) J Biomol Struct Dyn 7: 795-809
- PubMed: 2310515
- DOI: https://doi.org/10.1080/07391102.1990.10508524
- Primary Citation of Related Structures:
5ANA - PubMed Abstract:
The structure of the self-complementary octamer d(GTACGTAC) has been analyzed by a single crystal X-ray diffraction method at 2.25 A resolution. The crystallographic R factor was 0.184 for all 1233 reflections at this resolution. In spite of the alternating purine-pyrimidine sequence, d(GTACGTAC) adopts the A-form conformation rather than the left-handed Z-form. The average helix twist and the mean rise per base pair are 32.1 degrees and 3.18 A, respectively. The d(GTACGTAC) helix is characterized by a wide open major groove and small base-pair tilt (9.7 degrees). The partial unwinding of the helix is observed only at the central pyrimidine-purine C-G step, but not at the other pyrimidine-purine T-A steps. Based on this study and six other X-ray studies, we propose a hypothesis that the A-DNA's are always unwound approximately 10 degrees at the C-G steps. Significant differences in base-pair stacking modes are seen between the purine-pyrimidine step and the pyrimidine-purine step. All deoxyribose rings adopt the C3'-endo conformation. All backbone torsion angles fall into the range expected for the A-DNA form, except for the nucleotide G5, whose alpha and gamma torsion angles adopt the trans, trans conformation instead of the common gauche-, gauche+ conformation.
Organizational Affiliation:
Department of Chemistry, University of Kansas, Lawrence 66045.