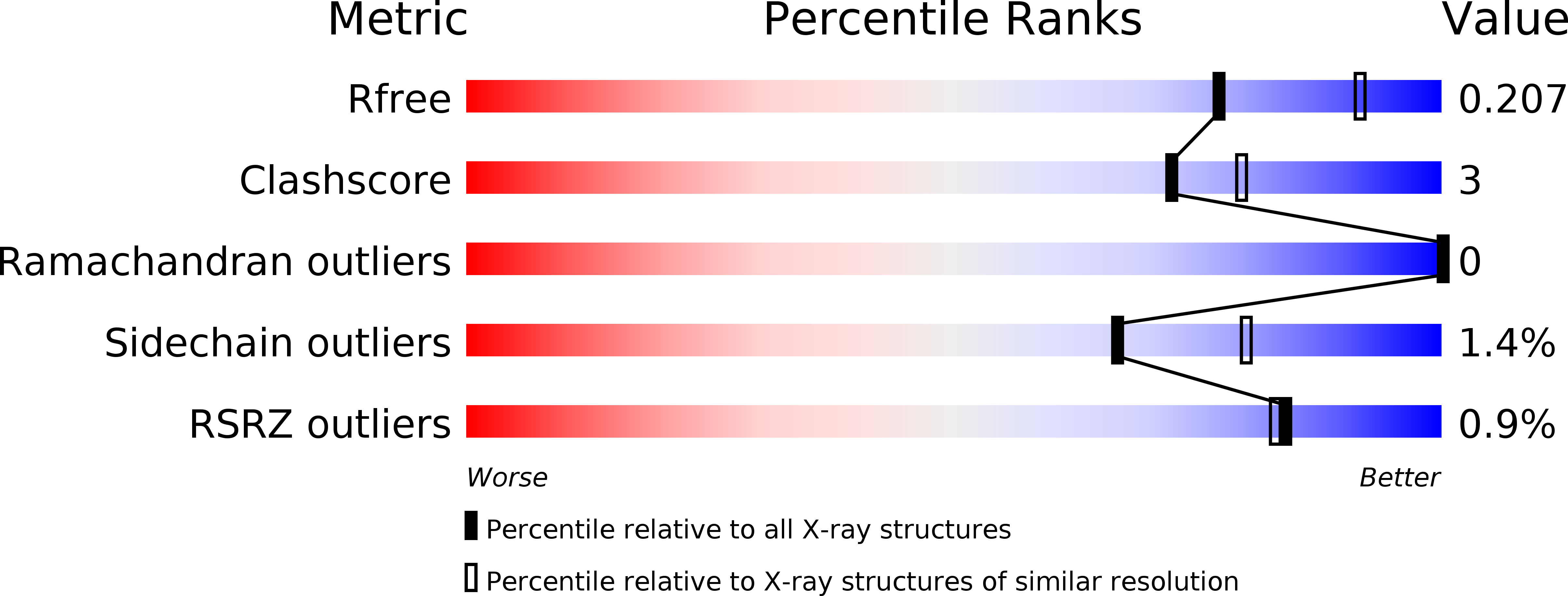
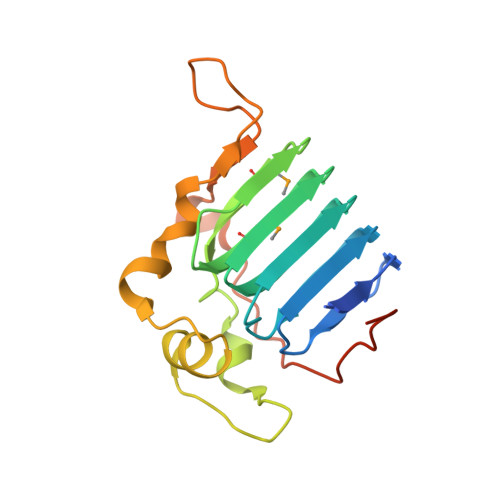
Crystal Structure of the C-Terminal Domain of Tubulin-Binding Cofactor C from Leishmania Major.
Barrack, K.L., Fyfe, P.K., Finney, A.J., Hunter, W.N.(2015) Mol Biochem Parasitol 201: 26
- PubMed: 25982270
- DOI: https://doi.org/10.1016/j.molbiopara.2015.05.003
- Primary Citation of Related Structures:
5AJ8 - PubMed Abstract:
Tubulin-binding cofactor C stimulates GTPase activity and contributes to the release of the heterodimeric α/β-tubulin from a super-complex of tubulin monomers and two ancillary cofactors. We have determined the 2.2 Å resolution crystal structure of the C-terminal domain of tubulin-binding cofactor C from Leishmania major based on single wavelength anomalous dispersion measurements targeting a selenomethionine derivative. Although previously predicted to consist of two domains the structure is best described as a single domain dominated by a right-handed β-helix of five turns that form a triangular prism. One face of the prism is covered by the C-terminal residues leaving another face solvent exposed. Comparisons with an orthologous human GTPase activating protein match key residues involved in binding nucleotide and identify the face of the β-helix fold likely involved in interacting with the β-tubulin:GTP complex.
Organizational Affiliation:
Division of Biological Chemistry & Drug Discovery, College of Life Sciences, University of Dundee, Dundee DD1 5EH, Scotland, UK.