Synthesis of Hydrolysis-Resistant Pyridoxal 5'-Phosphate Analogs and Their Biochemical and X-Ray Crystallographic Characterization with the Pyridoxal Phosphatase Chronophin.
Knobloch, G., Jabari, N., Stadlbauer, S., Schindelin, H., Kohn, M., Gohla, A.(2015) Bioorg Med Chem 23: 2819
- PubMed: 25783190
- DOI: https://doi.org/10.1016/j.bmc.2015.02.049
- Primary Citation of Related Structures:
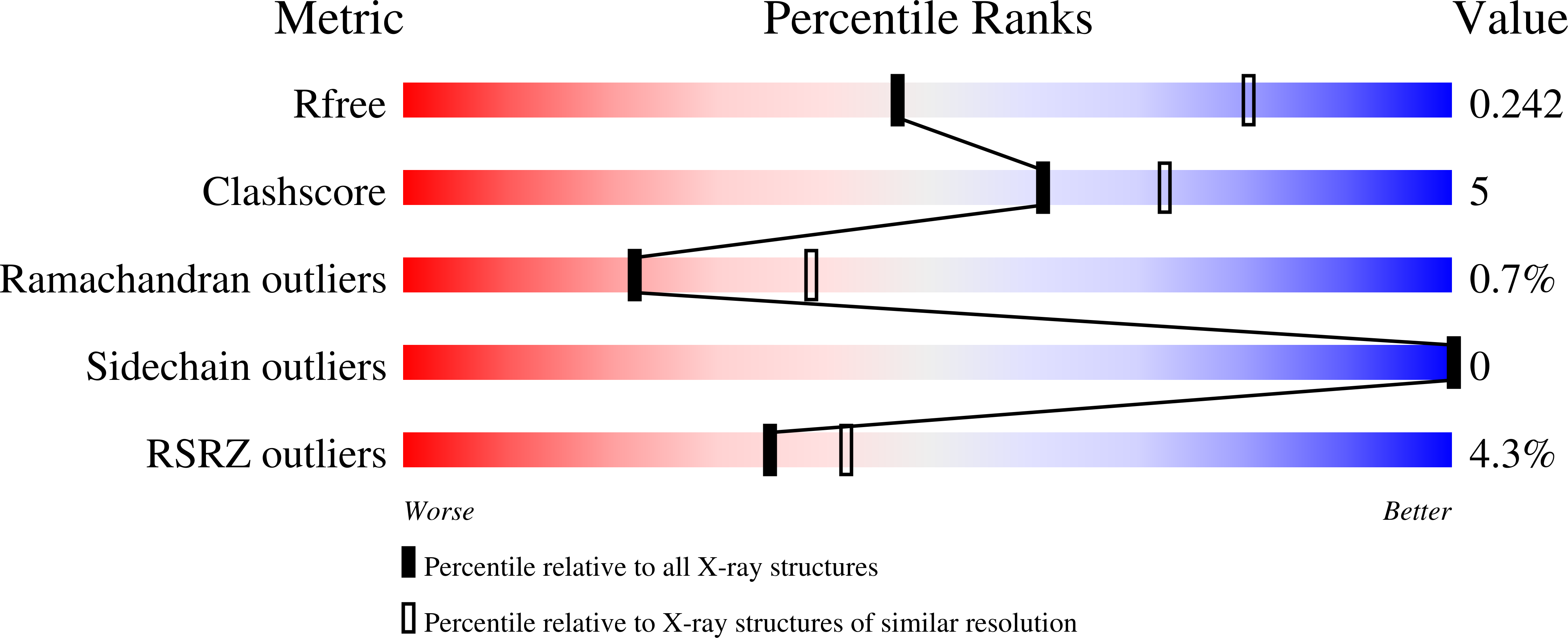
5AES - PubMed Abstract:
A set of phosphonic acid derivatives (1-4) of pyridoxal 5'-phosphate (PLP) was synthesized and characterized biochemically using purified murine pyridoxal phosphatase (PDXP), also known as chronophin. The most promising compound 1 displayed primarily competitive PDXP inhibitory activity with an IC50 value of 79μM, which was in the range of the Km of the physiological substrate PLP. We also report the X-ray crystal structure of PDXP bound to compound 3, which we solved to 2.75Å resolution (PDB code 5AES). The co-crystal structure proves that compound 3 binds in the same orientation as PLP, and confirms the mode of inhibition to be competitive. Thus, we identify compound 1 as a PDXP phosphatase inhibitor. Our results suggest a strategy to design new, potent and selective PDXP inhibitors, which may be useful to increase the sensitivity of tumor cells to treatment with cytotoxic agents.
Organizational Affiliation:
Institute of Pharmacology and Toxicology, University of Würzburg, Versbacher Strasse 9, D-97078 Würzburg, Germany; Rudolf Virchow Center for Experimental Biomedicine, University of Würzburg, Josef-Schneider-Strasse 2, D-97080 Würzburg, Germany.