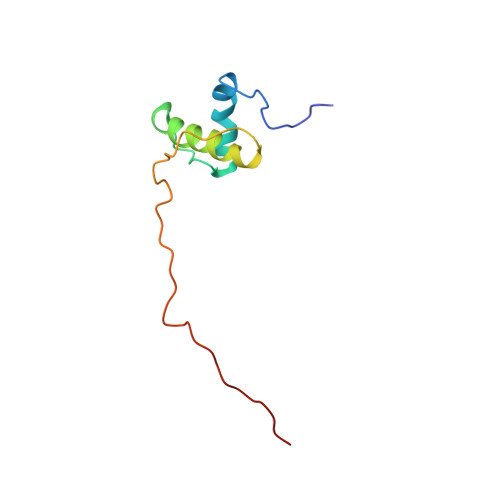
Structure of the N-Terminal Domain of the Metalloprotease Prtv from Vibrio Cholerae.
Edwin, A., Persson, C., Mayzel, M., Wai, S.N., Ohman, A., Karlsson, B.G., Sauer-Eriksson, A.E.(2015) Protein Sci 24: 2076
- PubMed: 26434928
- DOI: https://doi.org/10.1002/pro.2815
- Primary Citation of Related Structures:
5ABK - PubMed Abstract:
The metalloprotease PrtV from Vibrio cholerae serves an important function for the ability of bacteria to invade the mammalian host cell. The protein belongs to the family of M6 proteases, with a characteristic zinc ion in the catalytic active site. PrtV constitutes a 918 amino acids (102 kDa) multidomain pre-pro-protein that undergoes several N- and C-terminal modifications to form a catalytically active protease. We report here the NMR structure of the PrtV N-terminal domain (residues 23-103) that contains two short α-helices in a coiled coil motif. The helices are held together by a cluster of hydrophobic residues. Approximately 30 residues at the C-terminal end, which were predicted to form a third helical structure, are disordered. These residues are highly conserved within the genus Vibrio, which suggests that they might be functionally important.
Organizational Affiliation:
Department of Chemistry, Umeå University, Umeå, SE-901 87, Sweden.