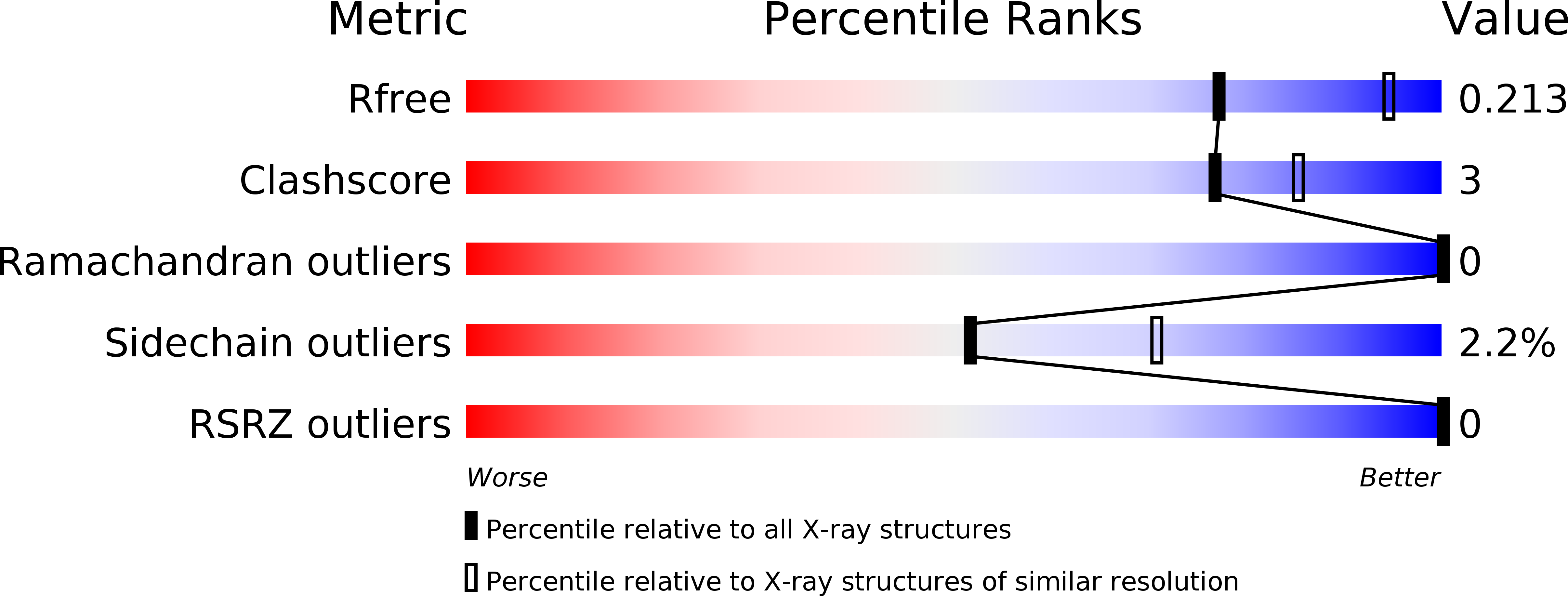
Crystal structure of the legume lectin-like domain of an ERGIC-53-like protein from Entamoeba histolytica
Khan, F., Suguna, K.(2019) Acta Crystallogr F Struct Biol Commun 75: 197-204
- PubMed: 30839295
- DOI: https://doi.org/10.1107/S2053230X19000499
- Primary Citation of Related Structures:
5ZBT - PubMed Abstract:
ERGIC-53-like proteins are type I membrane proteins that belong to the class of intracellular cargo receptors and are known to be indispensable for the intracellular transport of glycoproteins. They are implicated in transporting glycoproteins between the endoplasmic reticulum and the Golgi body. The crystal structure of the legume lectin-like domain of an ERGIC-53-like protein from Entamoeba histolytica has been determined at 2.4 Å resolution. Although the overall structure of the domain resembles those of its mammalian and yeast orthologs (ERGIC-53 and Emp46, respectively), there are significant changes in the carbohydrate-binding site. A sequence-based search revealed the presence of several homologs of ERGIC-53 in different species of Entamoeba. This is the first report of the structural characterization of a member of this class of proteins from a protozoan and serves to further knowledge and understanding regarding the species-specific differences.
Organizational Affiliation:
Molecular Biophysics Unit, Indian Institute of Science, Bangalore, Karnataka 560 012, India.