Structural basis underlying complex assembly and conformational transition of the type I R-M system.
Liu, Y.P., Tang, Q., Zhang, J.Z., Tian, L.F., Gao, P., Yan, X.X.(2017) Proc Natl Acad Sci U S A 114: 11151-11156
- PubMed: 28973912
- DOI: https://doi.org/10.1073/pnas.1711754114
- Primary Citation of Related Structures:
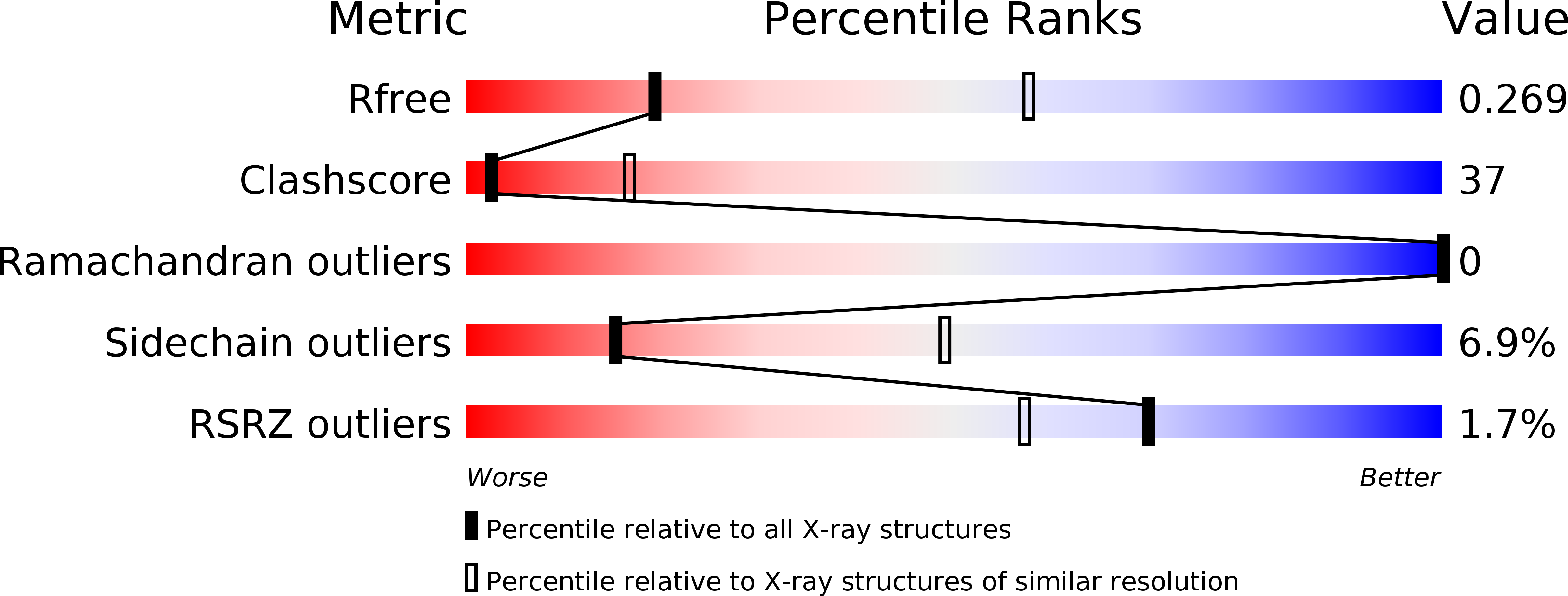
5YBB - PubMed Abstract:
Type I restriction-modification (R-M) systems are multisubunit enzymes with separate DNA-recognition (S), methylation (M), and restriction (R) subunits. Despite extensive studies spanning five decades, the detailed molecular mechanisms underlying subunit assembly and conformational transition are still unclear due to the lack of high-resolution structural information. Here, we report the atomic structure of a type I MTase complex (2M+1S) bound to DNA and cofactor S-adenosyl methionine in the "open" form. The intermolecular interactions between M and S subunits are mediated by a four-helix bundle motif, which also determines the specificity of the interaction. Structural comparison between open and previously reported low-resolution "closed" structures identifies the huge conformational changes within the MTase complex. Furthermore, biochemical results show that R subunits prefer to load onto the closed form MTase. Based on our results, we proposed an updated model for the complex assembly. The work reported here provides guidelines for future applications in molecular biology.
Organizational Affiliation:
National Laboratory of Biomacromelecules, Chinese Academy of Sciences (CAS) Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.