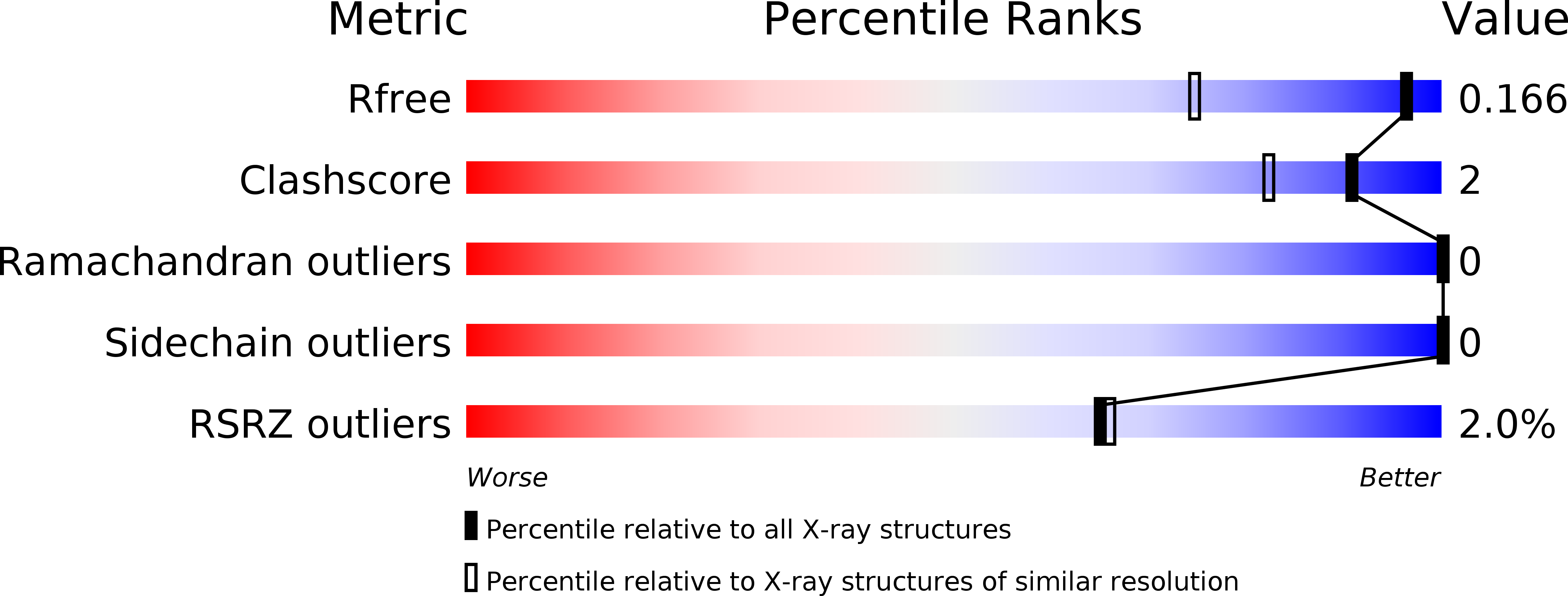
Structural mapping of hot spots within human CASPR2 discoidin domain for autoantibody recognition.
Liang, W., Zhang, J., Saint-Martin, M., Xu, F., Noraz, N., Liu, J., Honnorat, J., Liu, H.(2019) J Autoimmun 96: 168-177
- PubMed: 30337146
- DOI: https://doi.org/10.1016/j.jaut.2018.09.012
- Primary Citation of Related Structures:
5Y4M - PubMed Abstract:
Accumulating evidence has showed that anti-CASPR2 autoantibodies occur in a long list of neurological immune disorders including limbic encephalitis (LE). Belonging to the well-known neurexin superfamily, CASPR2 has been suggested to be a central node in the molecular networks controlling neurodevelopment. Distinct from other subfamilies in the neurexin superfamily, the CASPR subfamily features a unique discoidin (Disc) domain. As revealed by our and others' recent studies, CASPR2 Disc domain bears a major epitope for autoantibodies. However, structural information on CASPR2 recognition by autoantibodies has been lacking. Here, we report the crystal structure of human CASPR2 Disc domain at a high resolution of 1.31 Å, which is the first atomic-resolution structure of the CASPR subfamily members. The Disc domain adopts a total β structure and folds into a distorted jellyroll-like barrel with a conserved disulfide-bond interlocking its N- and C-termini. Defined by four loops and located in one end of the barrel, the "loop-tip surface" is totally polar and easily available for protein docking. Based on structure-guided epitope prediction, we generated nine mutants and evaluated their binding to autoantibodies of cerebrospinal fluid from twelve patients with limbic encephalitis. The quadruple mutant G69N/A71S/S77N/D78R impaired CASPR2 binding to autoantibodies from eleven LE patients, which indicates that the loop L1 in the Disc domain bears hot spots for autoantibody interaction. Structural mapping of autoepitopes within human CASPR2 Disc domain sheds light on how autoantibodies could sequester CASPR2 ectodomain and antagonize its functionalities in the pathogenic processes.
Organizational Affiliation:
State Key Laboratory of Natural and Biomimetic Drugs & School of Pharmaceutical Sciences, Peking University Health Science Center, 38 Xueyuan Road, Haidian District, Beijing 100191, China; Department of Molecular and Cellular Pharmacology, School of Pharmaceutical Sciences, Peking University Health Science Center, 38 Xueyuan Road, Haidian District, Beijing 100191, China.