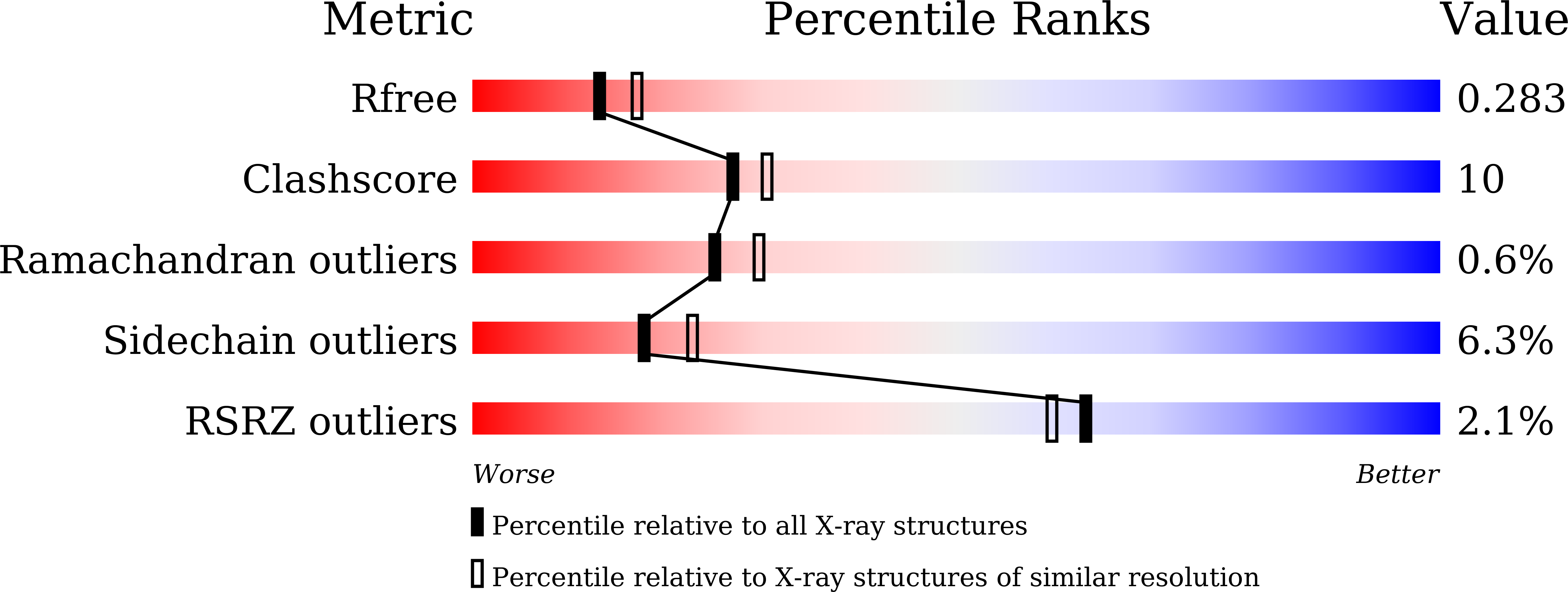
Fragment-Based Discovery of Pyrimido[1,2-b]indazole PDE10A Inhibitors.
Chino, A., Seo, R., Amano, Y., Namatame, I., Hamaguchi, W., Honbou, K., Mihara, T., Yamazaki, M., Tomishima, M., Masuda, N.(2018) Chem Pharm Bull (Tokyo) 66: 286-294
- PubMed: 29491261
- DOI: https://doi.org/10.1248/cpb.c17-00836
- Primary Citation of Related Structures:
5XUI, 5XUJ - PubMed Abstract:
In this study, we report the identification of potent pyrimidoindazoles as phosphodiesterase10A (PDE10A) inhibitors by using the method of fragment-based drug discovery (FBDD). The pyrazolopyridine derivative 2 was found to be a fragment hit compound which could occupy a part of the binding site of PDE10A enzyme by using the method of the X-ray co-crystal structure analysis. On the basis of the crystal structure of compound 2 and PDE10A protein, a number of compounds were synthesized and evaluated, by means of structure-activity relationship (SAR) studies, which culminated in the discovery of a novel pyrimidoindazole derivative 13 having good physicochemical properties.
Organizational Affiliation:
Drug Discovery Research, Astellas Pharma Inc.