Catalytic mechanism of glycoside hydrolase family 81 beta-1,3-glucanase
Yang, S., Qin, Z., Zhou, P., Yan, Q., Jiang, Z.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Endo-beta-1,3-glucanase | 796 | Rhizomucor miehei | Mutation(s): 0 EC: 3.2.1.39 | 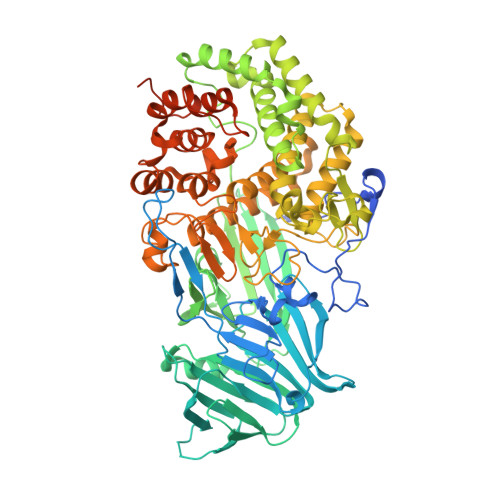 | |
UniProt | |||||
Find proteins for A0A023I7E1 (Rhizomucor miehei) Explore A0A023I7E1 Go to UniProtKB: A0A023I7E1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A023I7E1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | E | 6 |  | N/A | |
Glycosylation Resources | |||||
GlyTouCan: G41202OE GlyCosmos: G41202OE GlyGen: G41202OE | |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | F, M | 5 |  | N/A | |
Glycosylation Resources | |||||
GlyTouCan: G03330ED GlyCosmos: G03330ED | |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | G, L, O, P, Q | 4 |  | N/A | |
Glycosylation Resources | |||||
GlyTouCan: G87841AX GlyCosmos: G87841AX GlyGen: G87841AX | |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_900024 Query on PRD_900024 | J, K | beta-laminaribiose | Oligosaccharide / Antimicrobial |  | |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 108.7 | α = 90 |
| b = 99.02 | β = 111.35 |
| c = 138.43 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHENIX | phasing |