Disabling Cas9 by an anti-CRISPR DNA mimic.
Shin, J., Jiang, F., Liu, J.J., Bray, N.L., Rauch, B.J., Baik, S.H., Nogales, E., Bondy-Denomy, J., Corn, J.E., Doudna, J.A.(2017) Sci Adv 3: e1701620-e1701620
- PubMed: 28706995
- DOI: https://doi.org/10.1126/sciadv.1701620
- Primary Citation of Related Structures:
5VZL - PubMed Abstract:
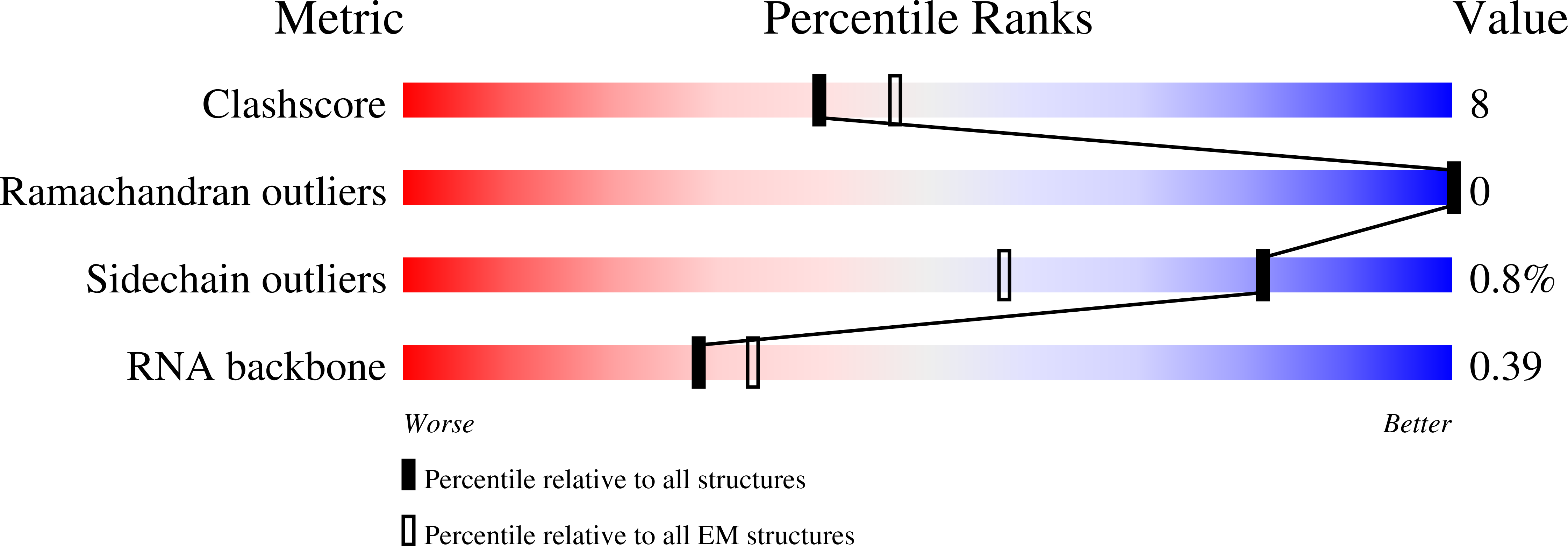
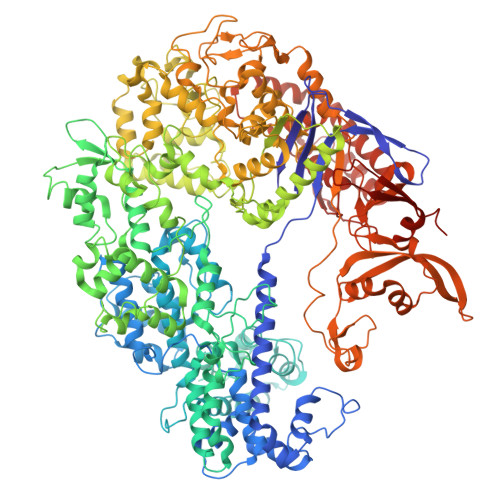
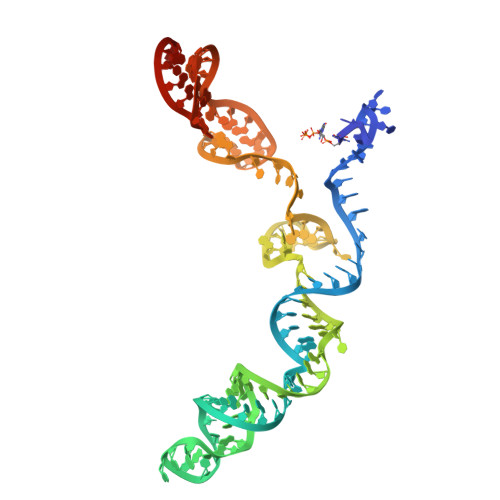
CRISPR (clustered regularly interspaced short palindromic repeats)-Cas9 gene editing technology is derived from a microbial adaptive immune system, where bacteriophages are often the intended target. Natural inhibitors of CRISPR-Cas9 enable phages to evade immunity and show promise in controlling Cas9-mediated gene editing in human cells. However, the mechanism of CRISPR-Cas9 inhibition is not known, and the potential applications for Cas9 inhibitor proteins in mammalian cells have not been fully established. We show that the anti-CRISPR protein AcrIIA4 binds only to assembled Cas9-single-guide RNA (sgRNA) complexes and not to Cas9 protein alone. A 3.9 Å resolution cryo-electron microscopy structure of the Cas9-sgRNA-AcrIIA4 complex revealed that the surface of AcrIIA4 is highly acidic and binds with a 1:1 stoichiometry to a region of Cas9 that normally engages the DNA protospacer adjacent motif. Consistent with this binding mode, order-of-addition experiments showed that AcrIIA4 interferes with DNA recognition but has no effect on preformed Cas9-sgRNA-DNA complexes. Timed delivery of AcrIIA4 into human cells as either protein or expression plasmid allows on-target Cas9-mediated gene editing while reducing off-target edits. These results provide a mechanistic understanding of AcrIIA4 function and demonstrate that inhibitors can modulate the extent and outcomes of Cas9-mediated gene editing.
Organizational Affiliation:
Innovative Genomics Institute, University of California, Berkeley, Berkeley, CA 94720, USA.