Generating enzyme and radical-mediated bisubstrates as tools for investigating Gcn5-related N-acetyltransferases.
Reidl, C., Majorek, K.A., Dang, J., Tran, D., Jew, K., Law, M., Payne, Y., Minor, W., Becker, D.P., Kuhn, M.L.(2017) FEBS Lett 591: 2348-2361
- PubMed: 28703494
- DOI: https://doi.org/10.1002/1873-3468.12753
- Primary Citation of Related Structures:
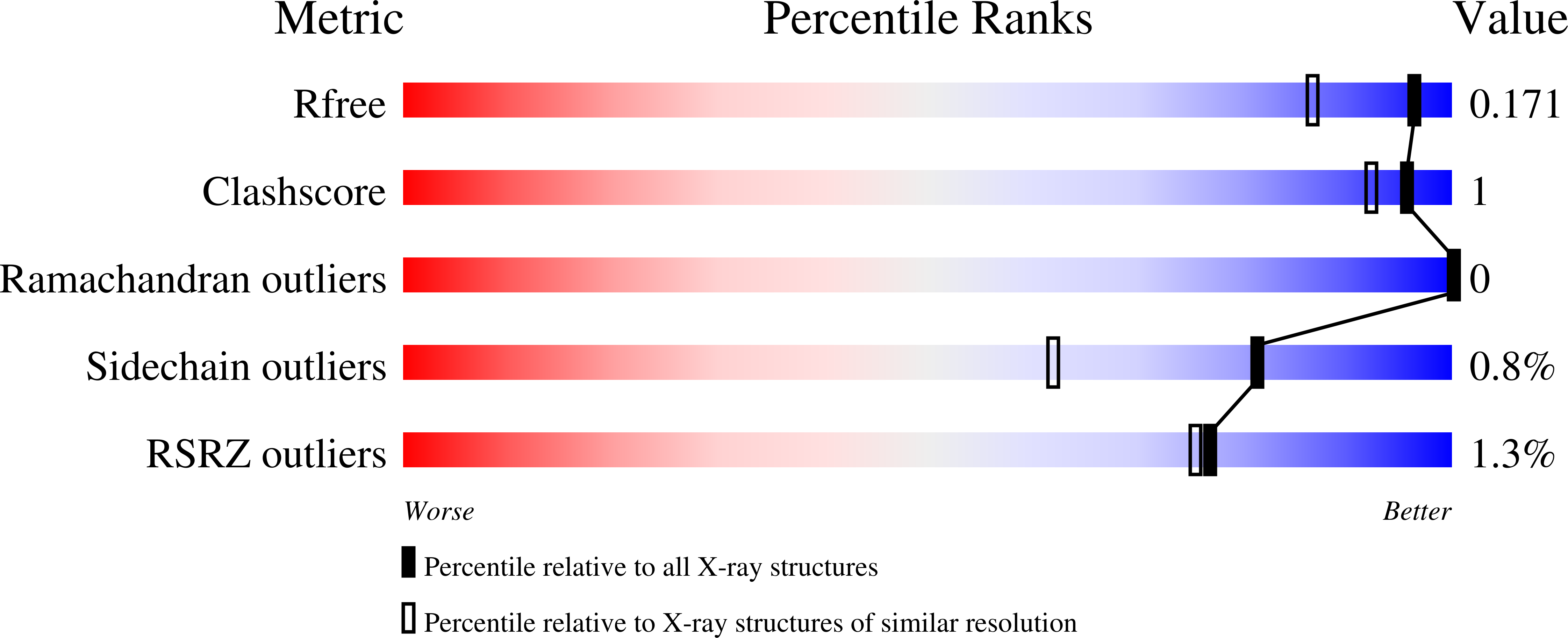
5VD6, 5VDB - PubMed Abstract:
Gcn5-related N-acetyltransferases (GNATs) are found in all kingdoms of life and catalyze important acyl transfer reactions in diverse cellular processes. While many 3D structures of GNATs have been determined, most do not contain acceptor substrates in their active sites. To expand upon existing crystallographic strategies for improving acceptor-bound GNAT structures, we synthesized peptide substrate analogs and reacted them with CoA in PA4794 protein crystals. We found two separate mechanisms for bisubstrate formation: (a) a novel X-ray induced radical-mediated alkylation of CoA with an alkene peptide and (b) direct alkylation of CoA with a halogenated peptide. Our approach is widely applicable across the GNAT superfamily and can be used to improve the success rate of obtaining liganded structures of other acyltransferases.
Organizational Affiliation:
Department of Chemistry, Loyola University Chicago, IL, USA.