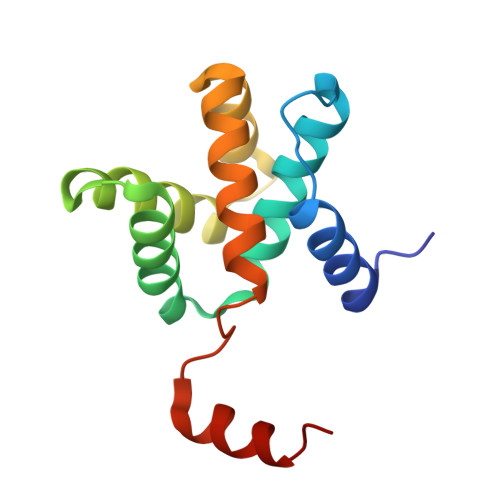
Ebola virus VP30 and nucleoprotein interactions modulate viral RNA synthesis.
Xu, W., Luthra, P., Wu, C., Batra, J., Leung, D.W., Basler, C.F., Amarasinghe, G.K.(2017) Nat Commun 8: 15576-15576
- PubMed: 28593988
- DOI: https://doi.org/10.1038/ncomms15576
- Primary Citation of Related Structures:
5VAO, 5VAP - PubMed Abstract:
Ebola virus (EBOV) is an enveloped negative-sense RNA virus that causes sporadic outbreaks with high case fatality rates. Ebola viral protein 30 (eVP30) plays a critical role in EBOV transcription initiation at the nucleoprotein (eNP) gene, with additional roles in the replication cycle such as viral assembly. However, the mechanistic basis for how eVP30 functions during the virus replication cycle is currently unclear. Here we define a key interaction between eVP30 and a peptide derived from eNP that is important to facilitate interactions leading to the recognition of the RNA template. We present crystal structures of the eVP30 C-terminus in complex with this eNP peptide. Functional analyses of the eVP30-eNP interface identify residues that are critical for viral RNA synthesis. Altogether, these results support a model where the eVP30-eNP interaction plays a critical role in transcription initiation and provides a novel target for the development of antiviral therapy.
Organizational Affiliation:
Department of Pathology and Immunology, Washington University School of Medicine, St Louis, Missouri 63105, USA.