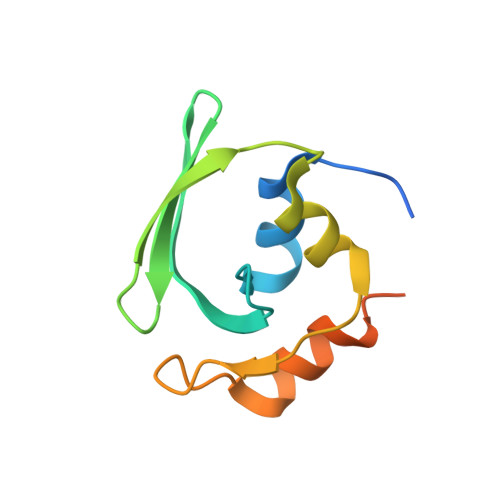
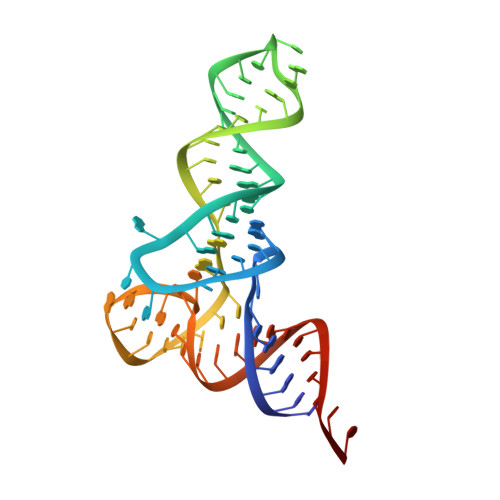Crystal structures reveal an elusive functional domain of pyrrolysyl-tRNA synthetase.
Suzuki, T., Miller, C., Guo, L.T., Ho, J.M.L., Bryson, D.I., Wang, Y.S., Liu, D.R., Soll, D.(2017) Nat Chem Biol 13: 1261-1266
- PubMed: 29035363
- DOI: https://doi.org/10.1038/nchembio.2497
- Primary Citation of Related Structures:
5UD5, 5V6X - PubMed Abstract:
Pyrrolysyl-tRNA synthetase (PylRS) is a major tool in genetic code expansion using noncanonical amino acids, yet its structure and function are not completely understood. Here we describe the crystal structure of the previously uncharacterized essential N-terminal domain of this unique enzyme in complex with tRNA Pyl . This structure explains why PylRS remains orthogonal in a broad range of organisms, from bacteria to humans. The structure also illustrates why tRNA Pyl recognition by PylRS is anticodon independent: the anticodon does not contact the enzyme. Then, using standard microbiological culture equipment, we established a new method for laboratory evolution-a noncontinuous counterpart of the previously developed phage-assisted continuous evolution. With this method, we evolved novel PylRS variants with enhanced activity and amino acid specificity. Finally, we employed an evolved PylRS variant to determine its N-terminal domain structure and show how its mutations improve PylRS activity in the genetic encoding of a noncanonical amino acid.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut, USA.