Heme Oxygenase 2 Binds Myristate to Regulate Retrovirus Assembly and TLR4 Signaling.
Zhu, Y., Luo, S., Sabo, Y., Wang, C., Tong, L., Goff, S.P.(2017) Cell Host Microbe 21: 220-230
- PubMed: 28132836
- DOI: https://doi.org/10.1016/j.chom.2017.01.002
- Primary Citation of Related Structures:
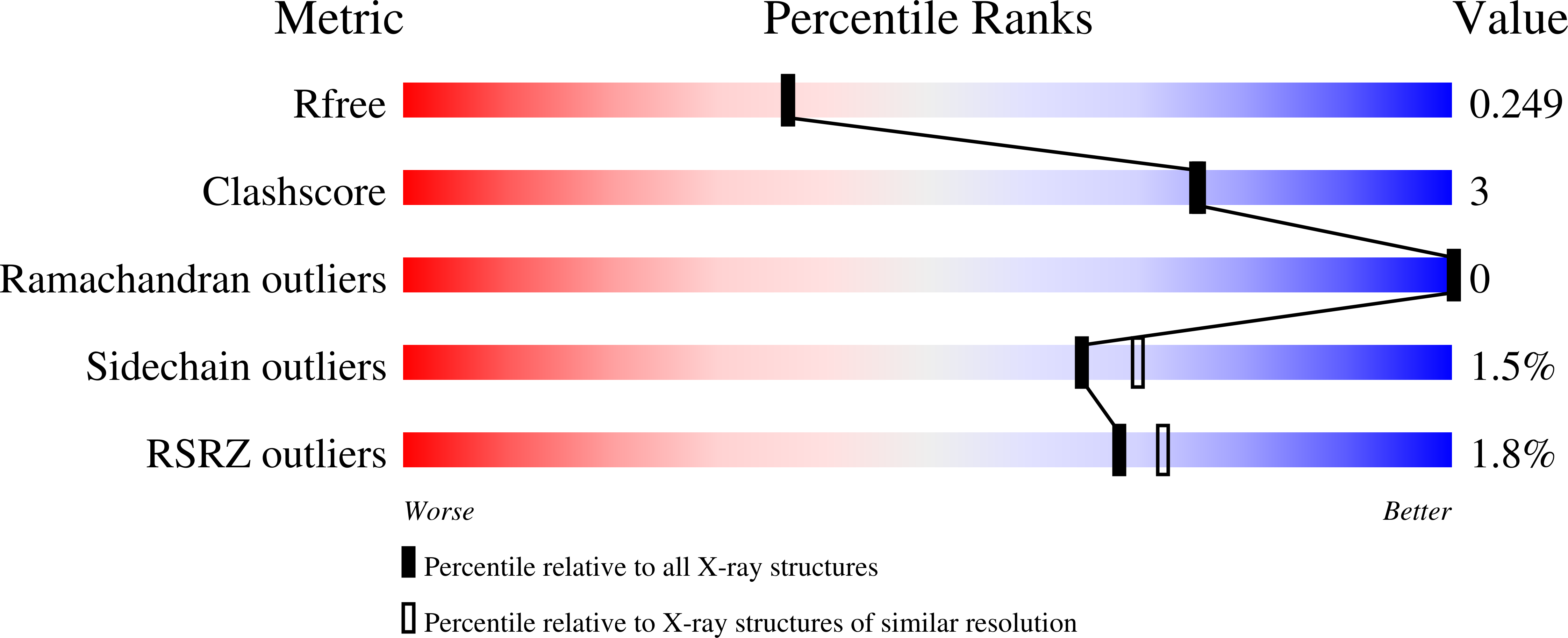
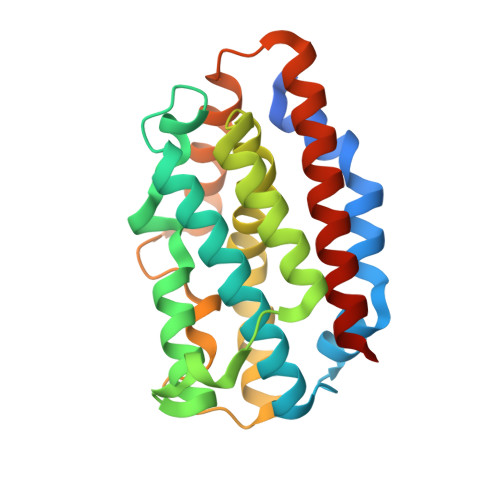
5UC8, 5UC9, 5UCA - PubMed Abstract:
N-myristoylation is the covalent attachment of myristic acid to the N terminus of proteins in eukaryotic cells. The matrix domain (MA) of HIV-1 Gag protein is N-myristoylated and plays an important role in virus budding. In screening for host factors that interact with HIV-1 MA, we found that heme oxygenase (HO-2) specifically binds the myristate moiety of Gag. HO-2 was also found to bind TRAM, an adaptor protein for Toll-like receptor 4 (TLR4), and thereby impact both virus replication and cellular inflammatory responses. A crystal structure revealed that HO-2 binds myristate via a hydrophobic channel adjacent to the heme-binding pocket. Inhibiting HO-2 expression, or blocking myristate binding with a heme analog, led to marked increases in virus production. HO-2 deficiency caused hyperresponsive TRAM-dependent TLR4 signaling and hypersensitivity to the TLR4 ligand lipopolysaccharide. Thus, HO-2 is a cellular myristate-binding protein that negatively regulates both virus replication and host inflammatory responses.
Organizational Affiliation:
Howard Hughes Medical Institute, Department of Biochemistry and Molecular Biophysics, and Department of Microbiology and Immunology, Columbia University, New York, NY 10032, USA.