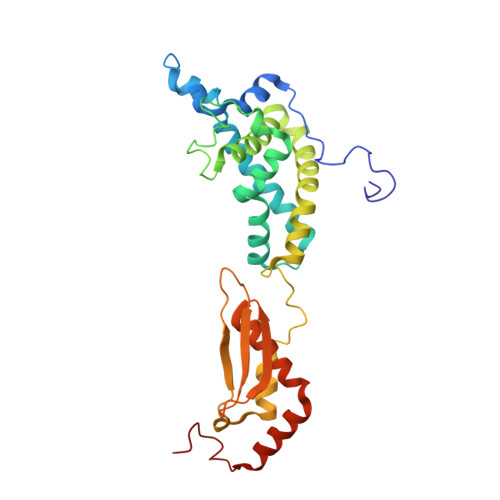
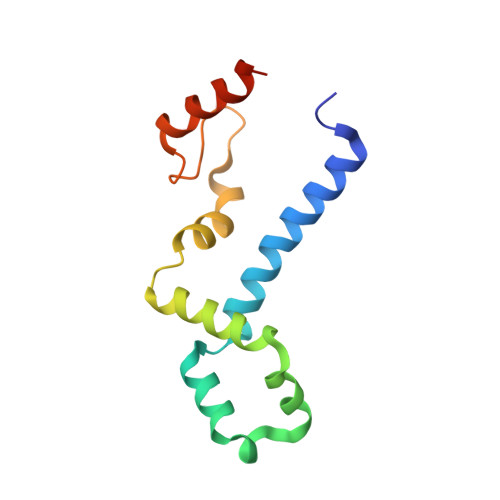
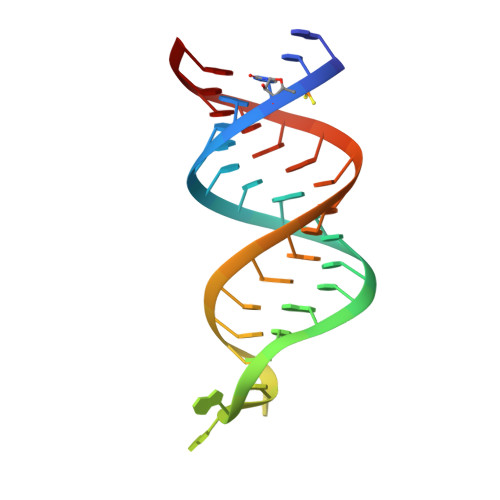
The Functional Cycle of Rnt1p: Five Consecutive Steps of Double-Stranded RNA Processing by a Eukaryotic RNase III.
Song, H., Fang, X., Jin, L., Shaw, G.X., Wang, Y.X., Ji, X.(2017) Structure 25: 353-363
- PubMed: 28111020
- DOI: https://doi.org/10.1016/j.str.2016.12.013
- Primary Citation of Related Structures:
5T16 - PubMed Abstract:
Double-stranded RNA (dsRNA)-specific RNase III proteins are required for RNA maturation and gene regulation. The mechanism of prokaryotic RNase IIIs has been well characterized, but how eukaryotic RNase IIIs (exemplified by Rnt1p, Drosha, and Dicer) work is less clear. Recently, we reported the crystal structure of Rnt1p in complex with RNA, revealing a double-ruler mechanism for substrate selection. Here, we present more structures of Rnt1p, either RNA free or RNA bound, featuring two major conformations of the enzyme. Using these structures with existing data, we describe the functional cycle of Rnt1p in five steps, selecting, loading, locking, cleavage, and releasing. We also describe atomic details of the two-Mg 2+ -ion catalytic mechanism that is applicable to all eukaryotic RNase III enzymes. Overall, our results indicate that substrate selection is achieved independent of cleavage, allowing the recognition of substrates with different structures while preserving the basic mechanism of cleavage.
Organizational Affiliation:
Macromolecular Crystallography Laboratory, National Cancer Institute, Frederick, MD 21702, USA.