Structure and dynamics of mesophilic variants from the homing endonuclease I-DmoI.
Alba, J., Marcaida, M.J., Prieto, J., Montoya, G., Molina, R., D'Abramo, M.(2017) J Comput Aided Mol Des 31: 1063-1072
- PubMed: 29177929
- DOI: https://doi.org/10.1007/s10822-017-0087-5
- Primary Citation of Related Structures:
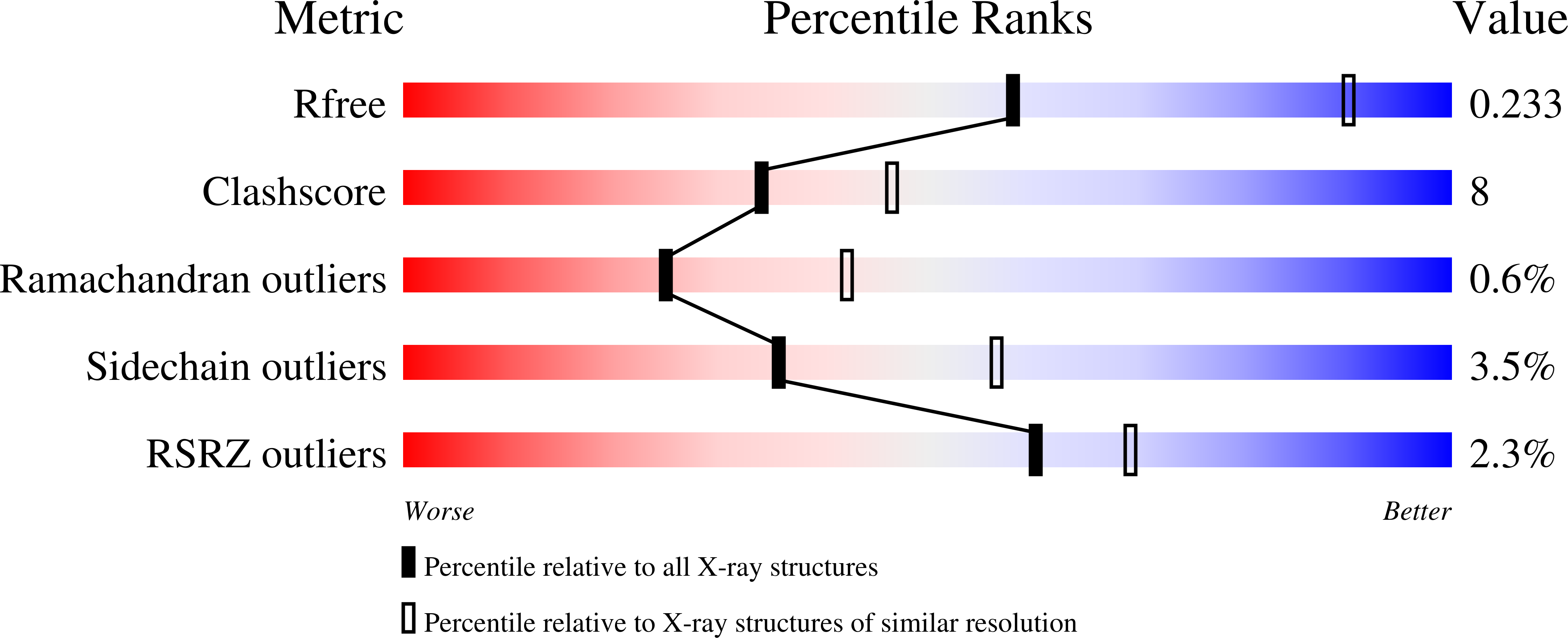
5O6G, 5O6I - PubMed Abstract:
I-DmoI, from the hyperthermophilic archaeon Desulfurococcus mobilis, belongs to the LAGLIDADG homing endonuclease protein family. Its members are highly specific enzymes capable of recognizing long DNA target sequences, thus providing potential tools for genome manipulation. Working towards this particular application, many efforts have been made to generate mesophilic variants of I-DmoI that function at lower temperatures than the wild-type. Here, we report a structural and computational analysis of two I-DmoI mesophilic mutants. Despite very limited structural variations between the crystal structures of these variants and the wild-type, a different dynamical behaviour near the cleavage sites is observed. In particular, both the dynamics of the water molecules and the protein perturbation effect on the cleavage site correlate well with the changes observed in the experimental enzymatic activity.
Organizational Affiliation:
Department of Chemistry, Sapienza University of Rome, P.le A. Moro, 5, 00185, Rome, Italy.