Functional and Structural Analysis of Programmed C-Methylation in the Biosynthesis of the Fungal Polyketide Citrinin.
Storm, P.A., Herbst, D.A., Maier, T., Townsend, C.A.(2017) Cell Chem Biol 24: 316-325
- PubMed: 28238725
- DOI: https://doi.org/10.1016/j.chembiol.2017.01.008
- Primary Citation of Related Structures:
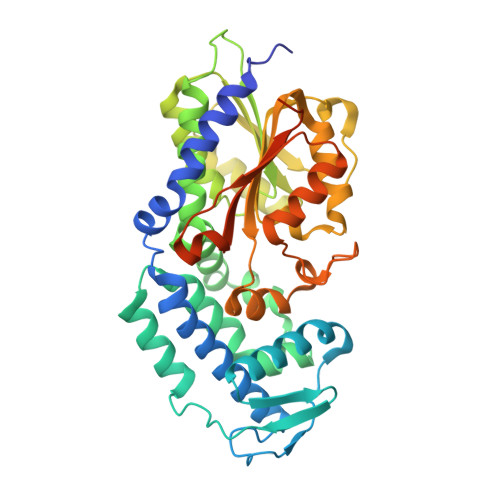
5MPT - PubMed Abstract:
Fungal polyketide synthases (PKSs) are large, multidomain enzymes that biosynthesize a wide range of natural products. A hallmark of these megasynthases is the iterative use of catalytic domains to extend and modify a series of enzyme-bound intermediates. A subset of these iterative PKSs (iPKSs) contains a C-methyltransferase (CMeT) domain that adds one or more S-adenosylmethionine (SAM)-derived methyl groups to the carbon framework. Neither the basis by which only specific positions on the growing intermediate are methylated ("programming") nor the mechanism of methylation are well understood. Domain dissection and reconstitution of PksCT, the fungal non-reducing PKS (NR-PKS) responsible for the first isolable intermediate in citrinin biosynthesis, demonstrates the role of CMeT-catalyzed methylation in precursor elongation and pentaketide formation. The crystal structure of the S-adenosyl-homocysteine (SAH) coproduct-bound PksCT CMeT domain reveals a two-subdomain organization with a novel N-terminal subdomain characteristic of PKS CMeT domains and provides insights into co-factor and ligand recognition.
Organizational Affiliation:
Department of Chemistry, Johns Hopkins University, Baltimore, MD 21218, USA.