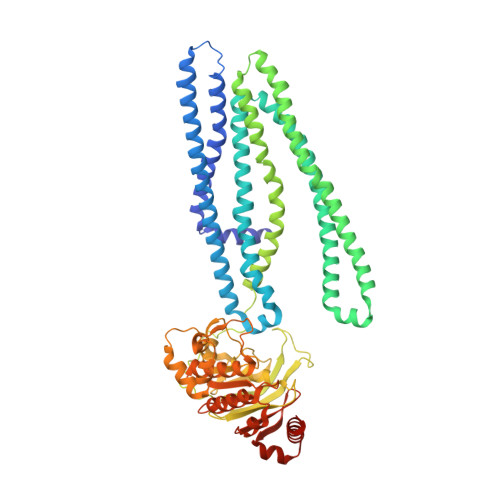
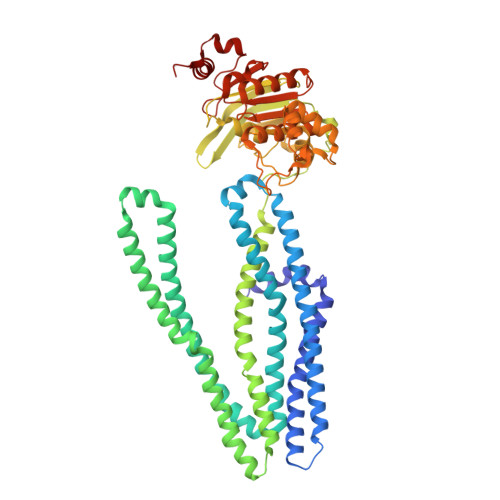
Crystal structure and mechanistic basis of a functional homolog of the antigen transporter TAP.
Noll, A., Thomas, C., Herbring, V., Zollmann, T., Barth, K., Mehdipour, A.R., Tomasiak, T.M., Bruchert, S., Joseph, B., Abele, R., Olieric, V., Wang, M., Diederichs, K., Hummer, G., Stroud, R.M., Pos, K.M., Tampe, R.(2017) Proc Natl Acad Sci U S A 114: E438-E447
- PubMed: 28069938
- DOI: https://doi.org/10.1073/pnas.1620009114
- Primary Citation of Related Structures:
5MKK - PubMed Abstract:
ABC transporters form one of the largest protein superfamilies in all domains of life, catalyzing the movement of diverse substrates across membranes. In this key position, ABC transporters can mediate multidrug resistance in cancer therapy and their dysfunction is linked to various diseases. Here, we describe the 2.7-Å X-ray structure of heterodimeric Thermus thermophilus multidrug resistance proteins A and B (TmrAB), which not only shares structural homology with the antigen translocation complex TAP, but is also able to restore antigen processing in human TAP-deficient cells. TmrAB exhibits a broad peptide specificity and can concentrate substrates several thousandfold, using only one single active ATP-binding site. In our structure, TmrAB adopts an asymmetric inward-facing state, and we show that the C-terminal helices, arranged in a zipper-like fashion, play a crucial role in guiding the conformational changes associated with substrate transport. In conclusion, TmrAB can be regarded as a model system for asymmetric ABC exporters in general, and for TAP in particular.
Organizational Affiliation:
Institute of Biochemistry, Biocenter, Goethe University Frankfurt, 60438 Frankfurt am Main, Germany.