Covalent Lectin Inhibition and Application in Bacterial Biofilm Imaging.
Wagner, S., Hauck, D., Hoffmann, M., Sommer, R., Joachim, I., Muller, R., Imberty, A., Varrot, A., Titz, A.(2017) Angew Chem Int Ed Engl 56: 16559-16564
- PubMed: 28960731
- DOI: https://doi.org/10.1002/anie.201709368
- Primary Citation of Related Structures:
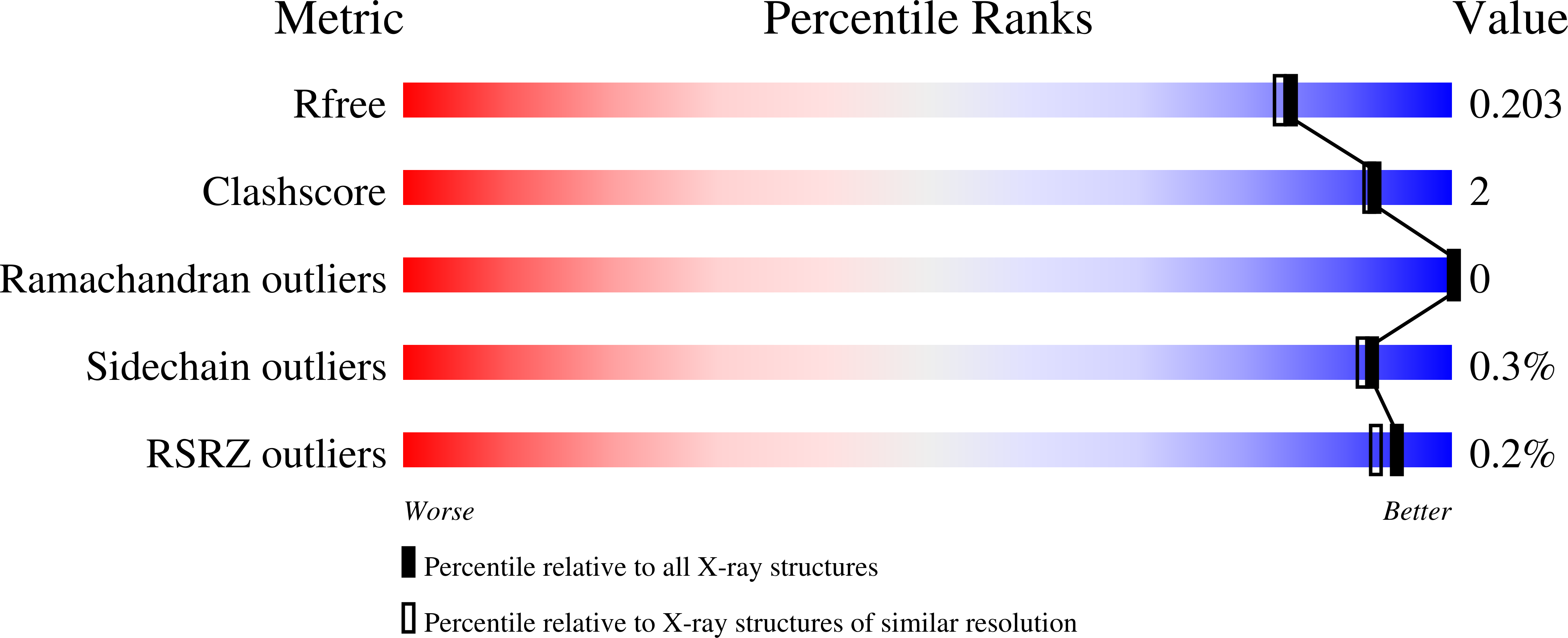
5MIH - PubMed Abstract:
Biofilm formation by pathogenic bacteria is a hallmark of chronic infections. In many cases, lectins play key roles in establishing biofilms. The pathogen Pseudomonas aeruginosa often exhibiting various drug resistances employs its lectins LecA and LecB as virulence factors and biofilm building blocks. Therefore, inhibition of the function of these proteins is thought to have potential in developing "pathoblockers" preventing biofilm formation and virulence. A covalent lectin inhibitor specific to a carbohydrate binding site is described for the first time. Its application in the LecA-specific in vitro imaging of biofilms formed by P. aeruginosa is also reported.
Organizational Affiliation:
Chemical Biology of Carbohydrates, Helmholtz Institute for Pharmaceutical Research Saarland (HIPS), 66123, Saarbrücken, Germany.