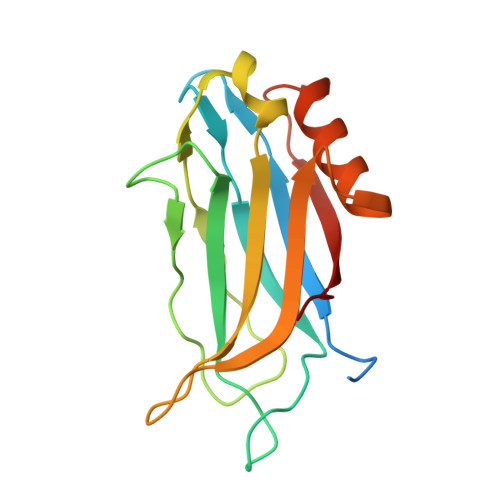
Structural characterization of the Rabphilin-3A-SNAP25 interaction.
Ferrer-Orta, C., Perez-Sanchez, M.D., Coronado-Parra, T., Silva, C., Lopez-Martinez, D., Baltanas-Copado, J., Gomez-Fernandez, J.C., Corbalan-Garcia, S., Verdaguer, N.(2017) Proc Natl Acad Sci U S A 114: E5343-E5351
- PubMed: 28634303
- DOI: https://doi.org/10.1073/pnas.1702542114
- Primary Citation of Related Structures:
5LO8, 5LOB, 5LOW - PubMed Abstract:
Membrane fusion is essential in a myriad of eukaryotic cell biological processes, including the synaptic transmission. Rabphilin-3A is a membrane trafficking protein involved in the calcium-dependent regulation of secretory vesicle exocytosis in neurons and neuroendocrine cells, but the underlying mechanism remains poorly understood. Here, we report the crystal structures and biochemical analyses of Rabphilin-3A C2B-SNAP25 and C2B-phosphatidylinositol 4,5-bisphosphate (PIP 2 ) complexes, revealing how Rabphilin-3A C2 domains operate in cooperation with PIP 2 /Ca 2+ and SNAP25 to bind the plasma membrane, adopting a conformation compatible to interact with the complete SNARE complex. Comparisons with the synaptotagmin1-SNARE show that both proteins contact the same SNAP25 surface, but Rabphilin-3A uses a unique structural element. Data obtained here suggest a model to explain the Ca 2+ -dependent fusion process by membrane bending with a myriad of variations depending on the properties of the C2 domain-bearing protein, shedding light to understand the fine-tuning control of the different vesicle fusion events.
Organizational Affiliation:
Structural Biology Unit, Institut de Biologia Molecular de Barcelona, Consejo Superior de Investigaciones Cientificas, 08028 Barcelona, Spain; cfocri@ibmb.csic.es senena@um.es nvmcri@ibmb.csic.es.