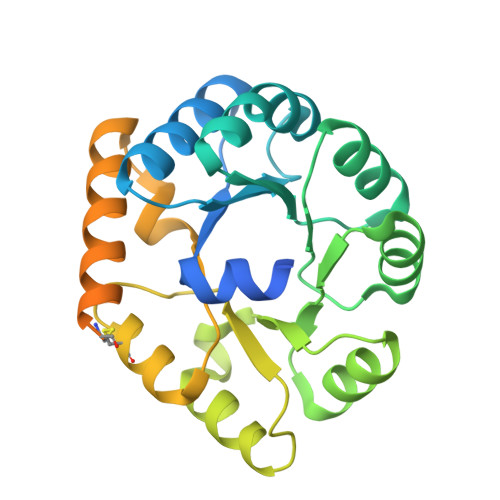
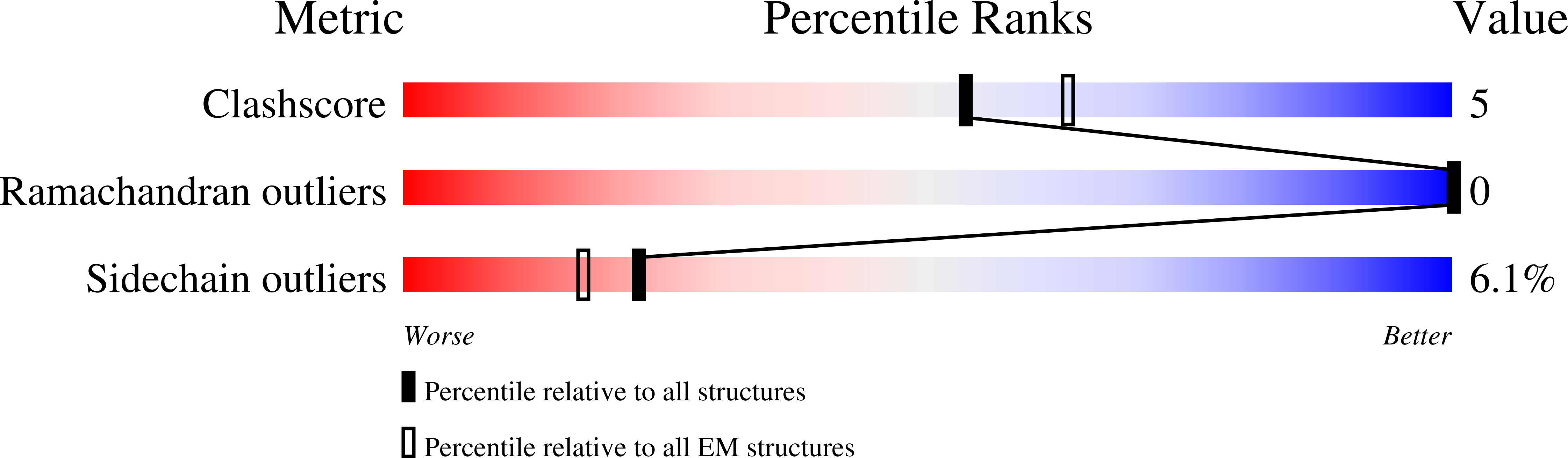Designed proteins induce the formation of nanocage-containing extracellular vesicles.
Votteler, J., Ogohara, C., Yi, S., Hsia, Y., Nattermann, U., Belnap, D.M., King, N.P., Sundquist, W.I.(2016) Nature 540: 292-295
- PubMed: 27919066
- DOI: https://doi.org/10.1038/nature20607
- Primary Citation of Related Structures:
5KP9 - PubMed Abstract:
Complex biological processes are often performed by self-organizing nanostructures comprising multiple classes of macromolecules, such as ribosomes (proteins and RNA) or enveloped viruses (proteins, nucleic acids and lipids). Approaches have been developed for designing self-assembling structures consisting of either nucleic acids or proteins, but strategies for engineering hybrid biological materials are only beginning to emerge. Here we describe the design of self-assembling protein nanocages that direct their own release from human cells inside small vesicles in a manner that resembles some viruses. We refer to these hybrid biomaterials as 'enveloped protein nanocages' (EPNs). Robust EPN biogenesis requires protein sequence elements that encode three distinct functions: membrane binding, self-assembly, and recruitment of the endosomal sorting complexes required for transport (ESCRT) machinery. A variety of synthetic proteins with these functional elements induce EPN biogenesis, highlighting the modularity and generality of the design strategy. Biochemical analyses and cryo-electron microscopy reveal that one design, EPN-01, comprises small (~100 nm) vesicles containing multiple protein nanocages that closely match the structure of the designed 60-subunit self-assembling scaffold. EPNs that incorporate the vesicular stomatitis viral glycoprotein can fuse with target cells and deliver their contents, thereby transferring cargoes from one cell to another. These results show how proteins can be programmed to direct the formation of hybrid biological materials that perform complex tasks, and establish EPNs as a class of designed, modular, genetically-encoded nanomaterials that can transfer molecules between cells.
Organizational Affiliation:
Department of Biochemistry, University of Utah, Salt Lake City, Utah 84112, USA.