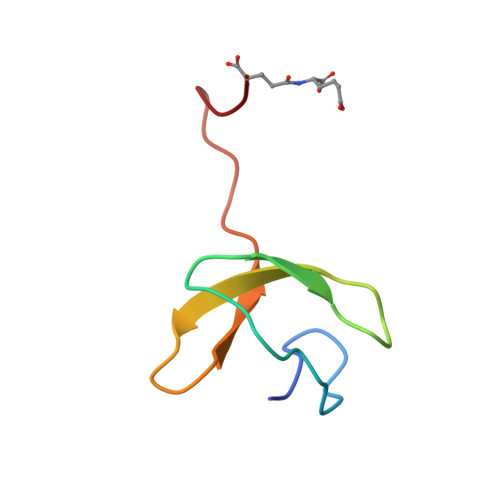
Lysine Biosynthesis of Thermococcus kodakarensis with the Capacity to Function as an Ornithine Biosynthetic System.
Yoshida, A., Tomita, T., Atomi, H., Kuzuyama, T., Nishiyama, M.(2016) J Biol Chem 291: 21630-21643
- PubMed: 27566549
- DOI: https://doi.org/10.1074/jbc.M116.743021
- Primary Citation of Related Structures:
5K2M - PubMed Abstract:
We recently discovered a biosynthetic system using a novel amino group carrier protein called LysW for lysine biosynthesis via α-aminoadipate (AAA), and revealed that this system is also utilized in the biosynthesis of arginine by Sulfolobus In the present study, we focused on the biosynthesis of lysine and ornithine in the hyperthermophilic archaeon Thermococcus kodakarensis, and showed that their biosynthesis is accomplished by a single set of metabolic enzymes. We also determined the crystal structure of the LysX family protein from T. kodakarensis, which catalyzes the conjugation of LysW with either AAA or glutamate, in a complex with LysW-γ-AAA. This crystal structure is the first example to show how LysX recognizes AAA as a substrate and provides a structural basis for the bifunctionality of the LysX family protein from T. kodakarensis Based on comparisons with other LysX family proteins, we propose a mechanism for substrate recognition and its relationship with molecular evolution among LysX family proteins, which have different substrate specificities.
Organizational Affiliation:
From the Biotechnology Research Center, University of Tokyo, 1-1-1, Yayoi, Bunkyo-ku, Tokyo 113-8657.