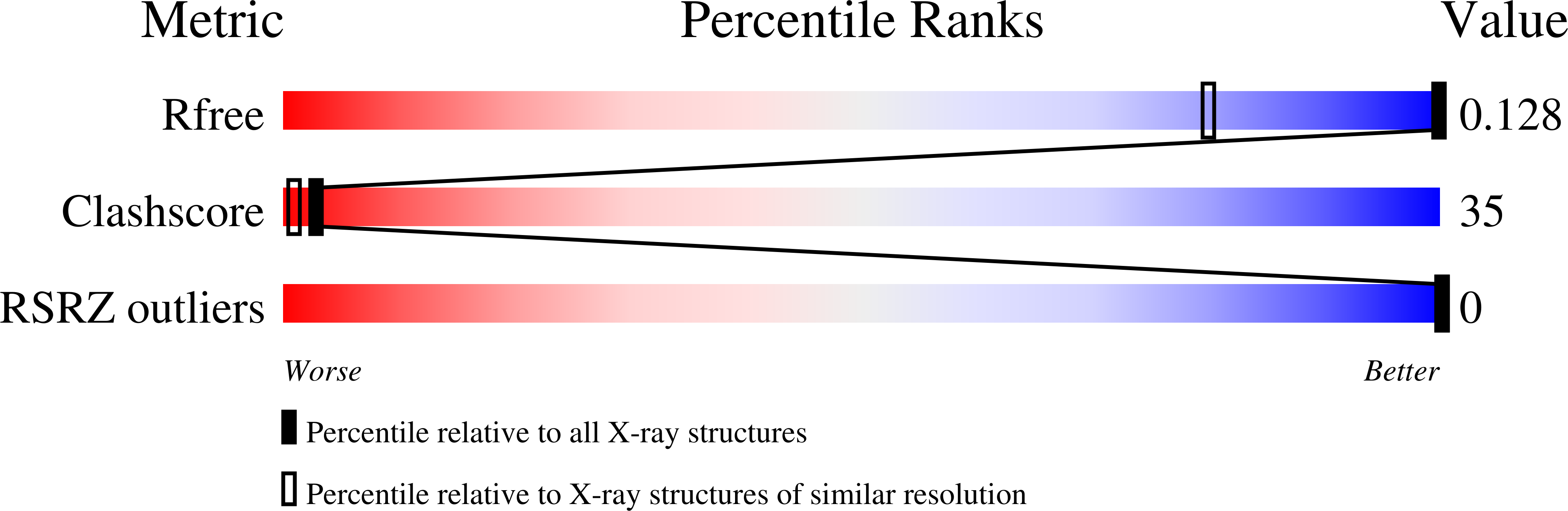Ultrahigh-resolution centrosymmetric crystal structure of Z-DNA reveals the massive presence of alternate conformations.
Drozdzal, P., Gilski, M., Jaskolski, M.(2016) Acta Crystallogr D Struct Biol 72: 1203-1211
- PubMed: 27841753
- DOI: https://doi.org/10.1107/S205979831601679X
- Primary Citation of Related Structures:
5JZQ - PubMed Abstract:
The self-complementary d(CGCGCG) hexanucleotide was synthesized with both D-2'-deoxyribose (the natural enantiomer) and L-2'-deoxyribose, and the two enantiomers were mixed in racemic (1:1) proportions and crystallized, producing a new crystal form with C2/c symmetry that diffracted X-rays to 0.78 Å resolution. The structure was solved by direct, dual-space and molecular-replacement methods and was refined to an R factor of 13.86%. The asymmetric unit of the crystal contains one Z-DNA duplex and three Mg 2+ sites. The crystal structure is comprised of both left-handed (D-form) and right-handed (L-form) Z-DNA duplexes and shows an unexpectedly high degree of structural disorder, which is manifested by the presence of alternate conformations along the DNA backbone chains as well as at four nucleobases (including one base pair) modelled in double conformations. The crystal packing of the presented D/L-DNA-Mg 2+ structure exhibits novel DNA hydration patterns and an unusual arrangement of the DNA helices in the unit cell. The paper describes the structure in detail, concentrating on the mode of disorder, and compares the crystal packing of the racemic d(CGCGCG) 2 duplex with those of other homochiral and heterochiral Z-DNA structures.
Organizational Affiliation:
Department of Crystallography, Faculty of Chemistry, A. Mickiewicz University, Poznan, Poland.